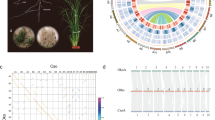
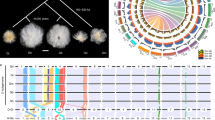
Abstract
Polyploidy and life-strategy transitions between annuality and perenniality often occur in flowering plants. However, the evolutionary propensities of polyploids and the genetic bases of such transitions remain elusive. We assembled chromosome-level genomes of representative perennial species across the genus Glycine including five diploids and a young allopolyploid, and constructed a Glycine super-pangenome framework by integrating 26 annual soybean genomes. These perennial diploids exhibit greater genome stability and possess fewer centromere repeats than the annuals. Biased subgenomic fractionation occurred in the allopolyploid, primarily by accumulation of small deletions in gene clusters through illegitimate recombination, which was associated with pre-existing local subgenomic differentiation. Two genes annotated to modulate vegetative–reproductive phase transition and lateral shoot outgrowth were postulated as candidates underlying the perenniality–annuality transition. Our study provides insights into polyploid genome evolution and lays a foundation for unleashing genetic potential from the perennial gene pool for soybean improvement.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Raw sequences generated during this study are deposited in the public repository of the National Center for Biotechnology Information under accession number PRJNA44023. The annotated assemblies are deposited in the European Nucleotide Archive under accession number PRJEB44023. Additionally, the assembled genome data and gene annotation have been deposited in SoyBase (http://soybase.org/data/v2/Glycine/) for future visualization of interspecific genome content comparison that is under development.
Code availability
The main custom scripts have been deposited in GitHub (https://github.com/Yongbinzhuang/Perennial_Soybean_Genome).
References
Sedivy, E. J., Wu, F. Q. & Hanzawa, Y. Soybean domestication: the origin, genetic architecture and molecular bases. New Phytol. 214, 539–553 (2017).
Sherman-Broyles, S., Bombarely, A., Grimwood, J., Schmutz, J. & Doyle, J. Complete plastome sequences from Glycine syndetika and six additional perennial wild relatives of soybean. G3 4, 2023–2033 (2014).
Schmutz, J. et al. Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183 (2010).
Li, Y. H. et al. De novoÿassembly of soybean wild relatives for pan-genome analysis of diversity and agronomic traits. Nat. Biotechnol. 32, 1045–1052 (2014).
Liu, Y. C. et al. Pan-genome of wild and cultivated soybeans. Cell 182, 162–176 (2020).
Hyten, D. L. et al. Impacts of genetic bottlenecks on soybean genome diversity. Proc. Natl Acad. Sci. USA 103, 16666–16671 (2006).
Koenen, E. J. M. et al. Large-scale genomic sequence data resolve the deepest divergences in the legume phylogeny and support a near-simultaneous evolutionary origin of all six subfamilies. New Phytol. 225, 1355–1369 (2020).
Doyle, J. J. & Egan, A. N. Dating the origins of polyploidy events. New Phytol. 186, 73–85 (2010).
Egan, A. N. & Doyle, J. J. A comparison of global, gene-specific, and relaxed clock methods in a comparative genomics framework: dating the polyploid history of soybean (Glycine max). Syst. Biol. 59, 534–547 (2010).
Bombarely, A., Coate, J. E. & Doyle, J. J. Mining transcriptomic data to study the origins and evolution of a plant allopolyploid complex. Peer J. 2, e391 (2014).
Simão, F. A. et al. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015).
Parra, G., Bradnam, K. & Korf, I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23, 1061–1067 (2007).
Xie, M. et al. A reference-grade wild soybean genome. Nat. Commun. 10, 1216 (2019).
Sherman-Broyles, S. et al. The wild side of a major crop: soybean’s perennial cousins from down under. Am. J. Bot. 101, 1651–1665 (2014).
Lavin, M., Herendeen, P. S. & Wojciechowski, M. F. Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Syst. Biol. 54, 575–594 (2005).
Ma, J. X. et al. Plant centromere organization: a dynamic structure with conserved functions. Trends Genet. 23, 134–139 (2007).
Comai, L., Maheshwari, S. & Marimuthu, P. A. Plant centromeres. Curr. Opin. Plant Biol. 36, 158–167 (2017).
Gill, N. et al. Molecular and chromosomal evidence for allopolyploidy in soybean. Plant Physiol. 151, 1167–1174 (2009).
Khan, A. W. et al. Super-pangenome by integrating the wild side of a species for accelerated crop improvement. Trends Plant Sci. 25, 148–158 (2020).
Friedman, J. The evolution of annual and perennial plant life histories: ecological correlates and genetic mechanisms. Annu. Rev. Ecol. Evol. Syst. 51, 461–481 (2020).
Wang, R. H. et al. PEP1 regulates perennial flowering in Arabis alpina. Nature 459, 423–427 (2009).
Hyun, Y. et al. A regulatory circuit conferring varied flowering response to cold in annual and perennial plants. Science 363, 409–412 (2019).
Yu, X. & Michaels, S. D. The Arabidopsis Paf1c complex component CDC73 participates in the modification of FLOWERING LOCUS C chromatin. Plant Physiol. 153, 1074–1084 (2010).
Chevalier, F. et al. Strigolactone promotes degradation of DWARF14, an α/β hydrolase essential for strigolactone signaling in Arabidopsis. Plant Cell 26, 1134–1150 (2014).
Arite, T. et al. d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol. 50, 1416–1424 (2009).
Wendel, J. F. The wondrous cycles of polyploidy in plants. Am. J. Bot. 102, 1753–1756 (2015).
Zhao, Meixia et al. Patterns and consequences of subgenome differentiation provide insights into the nature of paleopolyploidy in plants. Plant Cell 29, 2974–2994 (2017).
Steige, K. A. & Slotte, T. Genomic legacies of the progenitors and the evolutionary consequences of allopolyploidy. Curr. Opin. Plant Biol. 30, 88–93 (2016).
Wendel, J. F., Lisch, D., Hu, G. & Mason, A. S. The long and short of doubling down: polyploidy, epigenetics, and the temporal dynamics of genome fractionation. Curr. Opin. Genet. Dev. 49, 1–7 (2018).
Hurgobin, B. et al. Homoeologous exchange is a major cause of gene presence/absence variation in the amphidiploid Brassica napus. Plant Biotechnol. J. 16, 1265–1274 (2018).
Salmon, A. et al. Homoeologous nonreciprocal recombination in polyploid cotton. New Phytol. 186, 123–134 (2010).
Bertioli, DavidJ. et al. The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 51, 877–884 (2019).
Mason, A. S. & Wendel, J. F. Homoeologous exchanges, segmental allopolyploidy, and polyploid genome evolution. Front. Genet. 11, 1014 (2020).
Devos, K. M., Brown, J. K. & Bennetzen, J. L. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 12, 1075–1079 (2002).
Ehrlich, S. D. in Mobile DNA (eds Berg, D. E. & Howe, M. M.) 799–832 (American Society for Microbiology, 1989).
McClintock, B. The significances of responses of the genome to challenge. Science 226, 792–801 (1984).
Bzymek, M. & Lovett, S. T. Instability of repetitive DNA sequences: the role of replication in multiple mechanisms. Proc. Natl Acad. Sci. USA 98, 8319–8325 (2001).
Gaut, B. S. et al. Recombination: an underappreciated factor in the evolution of plant genomes. Nat. Rev. Genet. 8, 77–84 (2007).
Smith, S. A. & Donoghue, M. J. Rates of molecular evolution are linked to life history in flowering plants. Science 322, 86–89 (2008).
Talbert, P. B. & Henikoff, S. What makes a centromere? Exp. Cell. Res. 389, 111895 (2020).
Lee, H. R. et al. Chromatin immunoprecipitation cloning reveals rapid evolutionary patterns of centromeric DNA in Oryza species. Proc. Natl Acad. Sci. USA 102, 11793–11798 (2005).
Cheng, Z. K. et al. Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon. Plant Cell 14, 1691–1704 (2002).
Li, Y. J. et al. Centromeric DNA characterization in the model grass Brachypodium distachyon provides insights on the evolution of the genus. Plant J. 93, 1088–1101 (2018).
Yoo, M. J., Liu, X., Pires, J. C., Soltis, P. S. & Soltis, D. E. Nonadditive gene expression in polyploids. Annu. Rev. Genet. 48, 485–517 (2014).
Chalhoub, B. et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950 (2014).
Hu, Y. et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 51, 739–748 (2019).
Ilut, D. C. et al. A comparative transcriptomic study of an allotetraploid and its diploid progenitors illustrates the unique advantages and challenges of RNA-Seq in plant species. Am. J. Bot. 99, 383–396 (2012).
Powell, A. F. & Doyle, J. J. Non-additive transcriptomic responses to inoculation with rhizobia in a young allopolyploid compared with its diploid progenitors. Genes 8, 357 (2017).
Doyle, J. J. & Coate, J. E. Polyploidy, the nucleotype, and novelty: the impact of genome doubling on the biology of the cell. Int. J. Plant Sci. https://doi.org/10.1086/700636 (2019).
Murray, M. & Thompson, W. F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4326 (1980).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Ranallo-Benavidez, T. R., Jaron, K. S. & Schatz, M. C. GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat. Commun. 11, 1432 (2020).
Chin, C. S. et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 13, 1050–1054 (2016).
Chin, C. S. et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569 (2013).
Walker, B. J. et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 9, e112963 (2014).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://arxiv.org/abs/1303.3997v2 (2013).
Camacho, C. et al. BLAST.: architecture and applications. BMC Bioinformatics 10, 421–429 (2009).
Gurevich, A. et al. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075 (2013).
Ellinghaus, D., Kurtz, S. & Willhoeft, U. LTRharvÿest, an efficient and flexible software for de novoÿdetection of LTR retrotransposons. BMC Bioinformatics 9, 18 (2008).
Ou, S. & Jiang, N. LTR_FINDER_parallel: parallelization of LTR_FINDER enabling rapid identification of long terminal repeat retrotransposons. Mob. DNA 10, 48 (2019).
Jurka, J. et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 110, 462–467 (2005).
Wicker, T. et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 8, 973–982 (2007).
Miele, V., Penel, S. & Duret, L. Ultra-fast sequence clustering from similarity networks with SiLiX. BMC Bioinformatics 12, 116–124 (2011).
Mao, H. & Wang, H. SINE_scan: an efficient tool to discover short interspersed nuclear elements (SINEs) in large-scale genomic datasets. Bioinformatics 33, 743–745 (2017).
Xiong, W. W. et al. HelitronScanner uncovers a large overlooked cache of Helitron transposons in many plant genomes. Proc. Natl Acad. Sci. USA 111, 10263–10268 (2014).
Du, J. C. et al. Evolutionary conservation, diversity and specificity of LTR retrotransposons in flowering plants: insights from genome-wide analysis and multi-specific comparison. Plant J. 63, 584–598 (2010).
Kumar, S. et al. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 (2018).
Ma, J. & Jackson, S. A. Retrotransposon accumulation and satellite amplification mediated by segmental duplication facilitate centromere expansion in rice. Genome Res. 16, 251–259 (2006).
Benson, G. Tandem repeats finder: a program to analyse DNA sequences. Nucleic Acids Res. 27, 573–580 (1999).
Marcais, G. & Kingsford, C. Jellyfish: A Fast k-mer Counter (2012); https://raw.githubusercontent.com/gmarcais/Jellyfish/master/doc/jellyfish.pdf
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T. & Salzberg, S. L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667 (2016).
Li, W. & Godzik, A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006).
Campbell, M. S. et al. MAKER-P: a tool kit for the rapid creation, management, and quality control of plant genome annotations. Plant Physiol. 164, 513–524 (2014).
Tarailo-Graovac, M. & Chen, N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinformatics 25, 4.10.1–4.10.14 (2009).
Pruitt, K. D., Tatusova, T. & Maglott, D. R. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35, D61–D65 (2007).
Korf, I. Gene finding in nÿovel genomes. BMC Bioinformatics 5, 59 (2004).
Stanke, M. & Waack, S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics 19, ii215–ii225 (2003).
Vaidya, G., Lohman, D. J. & Meier, R. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27, 171–180 (2011).
Bouckaert, R. et al. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 10, e1003537 (2014).
Lemey, P., Rambaut, A., Welch, J. J. & Suchard, M. A. Phylogeography takes a relaxed random walk in continuous space and time. Mol. Biol. Evol. 27, 1877–1885 (2010).
Drummond, A. J. & Suchard, M. A. Bayesian random local clocks, or one rate to rule them all. BMC Biol. 8, 114 (2010).
Koenen, E. J. M. et al. The origin of the legumes is a complex paleopolyploid phylogenomic tangle closely associated with the Cretaceous–Paleogene (K–Pg) mass extinction event. Syst. Biol. 70, 508–526 (2021).
Rambaut, A., Drummond, A. J., Xie, D., Baele, G. & Suchard, M. A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 67, 901–904 (2018).
Brown, J. W. & Smith, S. A. The past sure is tense: on interpreting phylogenetic divergence time estimates. Syst. Biol. 67, 340–353 (2017).
Lavin, M., Herendeen, P. S. & Wojciechowski, M. F. Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lingeages during the tertiary. Syst. Biol. 54, 575–594 (2005).
Wang, Y. et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49 (2012).
Qiao, X. et al. Gene duplication and evolution in recurring polyploidization–diploidization cycles in plants. Genome Biol. 20, 38 (2019).
Zhang, Z. et al. KaKs_Calculÿator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics 4, 259–263 (2006).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Suyama, M., Torrents, D. & Bork, P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34, W609–W612 (2006).
Goel, M., Sun, H., Jiao, W. B. & Schneeberger, K. SyRI: finding genomic rearrangements and local sequence differences from whole-genome assemblies. Genome Biol. 20, 277 (2019).
Marcais, G. et al. MUMmer4: a fast and versatile genome alignment system. PLoS Comput. Biol. 14, e1005944 (2018).
McDonald, J. H. & Kreitman, M. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351, 652–654 (1991).
Egea, R., Casillas, S. & Barbadilla, A. Standard and generalized McDonald–Kreitman test: a website to detect selection by comparing different classes of DNA sites. Nucleic Acids Res. 36, W157–W162 (2008).
Zhou, Z. et al. Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat. Biotechnol. 33, 408–414 (2015).
Yang, J. et al. The I-TASSER suite: protein structure and function prediction. Nat. Methods 12, 7–8 (2014).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Acknowledgements
We thank J. Campbell for preparing BES data generated from the SoyMapII project and A. Farmer for integrating the genome sequence data generated in this study into SoyBase. The work was mainly supported by the National Key Research and Development Program (grant no. 2021YFF1001203), the Taishan Scholars Program of Shandong Province (tsqn201812036), the Agricultural Variety Improvement Project of Shandong Province (2019LZGC004) and the Program for Scientific Research Innovation Team of Young Scholar in Colleges and Universities of Shandong Province, China (2020KJF008) to D.Z., Y.Z. and X.S.Z. Partial support was provided by the US National Science Foundation Plant Genome Research Program to S.A.J., J.J.D., S.B.C., J.G., J.S. and J.M. (IOS-0822258) and the US Department of Agriculture (USDA) National Institute of Food and Agriculture MultiState Project to J.J.D. and J.B.L. (NC7 1014310). This research was also supported in part by the USDA Agricultural Research Service, project 5030-21000-069-00D to S.B.C. for integration of this source of genome data into SoyBase. The findings and conclusions in this publication are those of the authors and should not be construed to represent any official USDA or US government determination or policy. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA.
Author information
Authors and Affiliations
Contributions
Y.Z., D.Z. and J.M. conceived and designed the research. J.G. and J.S. generated BES data. Y.Z., X.W., X.L., J.H., L.F, J.B.L. and D.Z. performed analysis. Y.Z., X.W., S.B.C., S.A.J, J.J.D., X.S.Z., D.Z and J.M. interpreted the data. Y.Z., D.Z. J.J.D. and J.M. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Plants thanks Hon-Ming Lam, Sanwen Huang, Xuelu Wang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Genomic collinearity and rearrangements among the annual and perennial Glycine species.
a, Syntenic plots of perennial Glycine species. b, Illustration of an segmental inversion occurred in the annual Glycine lineage. c, Illustration of chromosomal rearrangements involving chromosomes 8 and 10 of the A, D, and F genomes. Red vertical bar indicates gaps in assemblies. d, Portion of chromosomes show conserved synteny through the whole chromosome in each perennial Glycine species. e, Portion of identified genome variations supported by species-specific BAC end sequences.
Extended Data Fig. 2 Comparison of LTR-retrotransposons in the annual and perennial Glycine species.
a, Ratios of solo LTRs to intact LTR-retrotransposons identified in each genome. b, c, Relative abundances and insertion times of LTR-RTs belonging to the five largest families in G. cyrtoloba (b) and G. falcata (c). A mutation rate of 1.3×10-8 per nucleotide per year was used for estimation of insertion times of individual LTR-RTs. d, e, Relative abundances of LTR-RTs belonging to the five most abundant copia-LTR-RT families (d) and the five most abundant gypsy-LTR-RT families (e) in the G. max and each perennial genome.
Extended Data Fig. 3 Comparison of centromeric satellite repeat families, Gm-Cent1, Gm-Cent2, and Gf-Cent.
a, Consensus sequence of Gf-Cent constructed based on 500 Gf-Cent repeats randomly selected from the F genome, the orange bars indicate highly variable nucleotide positions (<0.7). b, Alignment of representative Gm-Cent1, Gm-Cent2 and Gf-Cent repeats.
Extended Data Fig. 4 Analysis of centromeric satellite repeats and their associated centromeric retrotransposon families in the G. max and perennial Glycine species.
a, Estimation of the divergent times of Gm-Cent1, Gm-Cent2, and Gf-Cent repeats. A mutation rate of 1.3×10-8 per nucleotide per year was used for estimation of divergence times. b, c, d, Relative abundance of putative centromere retrotransposon families with detected internal sequences, which were predicted to encode five key enzymes (GAP, AP, INT, RT and RNase H).
Extended Data Fig. 5 Relative abundances of singletons and duplicated genes in G. max and the perennial Glycine genomes.
a, The numbers of core genes identified in 26 annual Glycine datasets. b, Boxplot showing the number of Non-core, core genes and Non-redundant genes for five random selected annual soybeans from 26 annual Glycine datasets. d, Bar plot showing the number of shared singletons c and duplicated genes d between G. max and individual perennial genomes. e, Proportions of singletons and duplicates identified in individual genomes. Undefined categories of genes such as tandem duplicates were not shown in the plots.
Extended Data Fig. 6
GO enrichment analysis of Glycine orthologs underwent adaptive evolution between the two subgenera.
Extended Data Fig. 7 Neighbour-joining tree of young (<0.35 MY) copia-LTR-RTs in the two subgenomes of the recent allopolyploid.
Highlighted elements represent nearly identical elements amplified from one of the two subgenomes and inserted into the other subgenome. after the recent allopolyploidy event.
Extended Data Fig. 8 Neighbour-joining tree of young (<0.35 MY) gypsy-LTR-RTs in the two subgenomes of the recent allopolyploid.
Highlighted elements represent nearly identical elements amplified from one of the two subgenomes and inserted into the other subgenome after the recent allopolyploidy event.
Extended Data Fig. 9 Neighbour-joining tree of young (<0.35 MY) copia-LTR-RTs from in the A and D genomes.
No nearly identical elements in A and D were found.
Extended Data Fig. 10 Neighbour-joining tree of young (<0.35 MY) gypsy-LTR-RTs from in the A and D genomes.
No nearly identical elements in A and D were found.
Supplementary information
Supplementary Information
Supplementary Figs. 1–8 and additional Fig. 1.
Supplementary Tables
Supplementary Tables 1–18: 1, Evaluation and correction of the raw assembled genomes using bacterial artificial chromosome sequences (BESs); 2, k-mer analysis of perennial Glycine genomes; 3, Statistics of the six assembled perennial Glycine genomes; 4, Genome assembly and annotation completeness evaluated by BUSCO; 5, Genome assembly and annotation completeness evaluated by CEGMA; 6, Numbers of annotated genes and TE content in the assembled perennial genomes; 7, Genomic rearrangements identified in the sequenced Glycine species using common bean as a reference; 8, PCR primers used in the validation of ten selected genome conversions; 9, Summary of copia-LTR-retrotransposons identified in the perennial Glycine genomes; 10, Summary of gypsy-LTR-retrotransposons identified in perennial Glycine species; 11, The top five most abundant tandem repeats in perennial Glycine species; 12, Synteny table among 26 annual and 5 perennial soybeans; 13, Duplicates and singletons in the annual and perennial Glycine genomes; 14, Classification of gene status; 15, List of genes showing adaptive evolution between annual and perennial soybeans; 16, McDonald–Kreitman tests for a single flowering controlling gene showing adaptive evolution as candidates underlying the life-strategy transition in Glycine; 17, List of genes used for analysis of subgenome fractionation in G. dolichocarpa; 18, Losses of genes in At and Dt that are orthologues to the core or non-core Glycine genes in A and D.
Supplementary Data
Genome coordinations of transposons identified in each perennial Glycine species.
Rights and permissions
About this article
Cite this article
Zhuang, Y., Wang, X., Li, X. et al. Phylogenomics of the genus Glycine sheds light on polyploid evolution and life-strategy transition. Nat. Plants 8, 233–244 (2022). https://doi.org/10.1038/s41477-022-01102-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-022-01102-4
This article is cited by
-
Pangenome analysis reveals transposon-driven genome evolution in cotton
BMC Biology (2024)
-
A sequence-aware merger of genomic structural variations at population scale
Nature Communications (2024)
-
Plant pangenomes for crop improvement, biodiversity and evolution
Nature Reviews Genetics (2024)
-
Technology-enabled great leap in deciphering plant genomes
Nature Plants (2024)
-
Discovery and genome-wide characterization of a novel miniature inverted repeat transposable element reveal genome-specific distribution in Glycine
Genes & Genomics (2024)