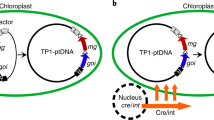
Abstract
Plant molecular farming, that is, using plants as hosts for production of therapeutic proteins and high-value compounds, has gained substantial interest in recent years. Chloroplasts in particular are an attractive subcellular compartment for expression of foreign genes. Here, we present a new method for transgene introduction and expression in chloroplasts that, unlike classically used approaches, does not require transgene insertion into the chloroplast genome. Instead, the transgene is amplified as a physically independent entity termed a ‘minichromosome’. Amplification occurs in the presence of a helper protein that initiates the replication process via recognition of specific sequences flanking the transgene, resulting in accumulation of extremely high levels of transgene DNA. Importantly, we demonstrate that such amplified transgenes serve as a template for foreign protein expression, are maintained stably during plant development and are maternally transmitted to the progeny. These findings indicate that the minichromosome-based approach is an attractive tool for transgene expression in chloroplasts and for organelle genome engineering.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Nucleotide sequences of the transformation cassettes are available under GenBank accessions MZ074313 (AJTB), MZ074315 (AJTE), MZ074314 (AJTD), MZ074316 (AJTG), MZ074319 (AJWY), MZ074310 (AIBW), MZ074312 (AIIS), MZ074311 (AIDR), MZ074318 (AJWU nRep) and MZ074317 (AJWO ptRep). Transformation vectors and transgenic plants will be made available on the execution of a Material Transfer Agreement with Algentech. The data supporting the findings of this study are presented within the article, Extended Data and Supplementary Information. Source data are provided with this paper.
References
Donini, M. & Marusic, C. Current state-of-the-art in plant-based antibody production systems. Biotechnol. Lett. 41, 335–346 (2019).
Kopertekh, L. & Schiemann, J. Transient production of recombinant pharmaceutical proteins in plants: evolution and perspectives. Curr. Med. Chem. 26, 365–380 (2017).
Holtz, B. R. et al. Commercial-scale biotherapeutics manufacturing facility for plant-made pharmaceuticals. Plant Biotechnol. J. 13, 1180–1190 (2015).
Tekoah, Y. et al. Large-scale production of pharmaceutical proteins in plant cell culture—the protalix experience. Plant Biotechnol. J. 13, 1199–1208 (2015).
Vamvaka, E. et al. Unexpected synergistic HIV neutralization by a triple microbicide produced in rice endosperm. Proc. Natl Acad. Sci. USA 115, E7854–E7862 (2018).
Margolin, E., Chapman, R., Williamson, A. L., Rybicki, E. P. & Meyers, A. E. Production of complex viral glycoproteins in plants as vaccine immunogens. Plant Biotechnol. J. 16, 1531–1545 (2018).
Fuentes, P., Armarego-Marriott, T. & Bock, R. Plastid transformation and its application in metabolic engineering. Curr. Opin. Biotechnol. 49, 10–15 (2018).
Bock, R. Engineering plastid genomes: methods, tools, and applications in basic research and biotechnology. Annu. Rev. Plant Biol. 66, 211–241 (2015).
Zhang, B., Shanmugaraj, B. & Daniell, H. Expression and functional evaluation of biopharmaceuticals made in plant chloroplasts. Curr. Opin. Chem. Biol. 38, 17–23 (2017).
Kuroda, H. & Maliga, P. Complementarity of the 16S rRNA penultimate stem with sequences downstream of the AUG destabilizes the plastid mRNAs. Nucleic Acids Res. 29, 970–975 (2001).
Ruf, S., Karcher, D. & Bock, R. Determining the transgene containment level provided by chloroplast transformation. Proc. Natl Acad. Sci. USA 104, 6998–7002 (2007).
Bohmert-Tatarev, K., McAvoy, S., Daughtry, S., Peoples, O. P. & Snell, K. D. High levels of bioplastic are produced in fertile transplastomic tobacco plants engineered with a synthetic operon for the production of polyhydroxybutyrate. Plant Physiol. 155, 1690–1708 (2011).
Legen, J. et al. Stabilization and translation of synthetic operon-derived mRNAs in chloroplasts by sequences representing PPR protein-binding sites. Plant J. 94, 8–21 (2018).
Lu, Y., Rijzaani, H., Karcher, D., Ruf, S. & Bock, R. Efficient metabolic pathway engineering in transgenic tobacco and tomato plastids with synthetic multigene operons. Proc. Natl Acad. Sci. USA 110, E623–E632 (2013).
Quesada-Vargas, T., Ruiz, O. N. & Daniell, H. Characterization of heterologous multigene operons in transgenic chloroplasts. Transcription, processing, and translation. Plant Physiol. 138, 1746–1762 (2005).
Long, B. M. et al. Carboxysome encapsulation of the CO2-fixing enzyme Rubisco in tobacco chloroplasts. Nat. Commun. 9, 3570 (2018).
Bock, R. Genetic engineering of the chloroplast: novel tools and new applications. Curr. Opin. Biotechnol. 26, 7–13 (2014).
Gleba, Y. Y., Tusé, D. & Giritch, A. in Plant Viral Vectors. Current Topics in Microbiology and Immunology Vol. 375 (eds Palmer K. & Gleba Y.) 155–192 (Springer, 2014).
Lozano-Durán, R. Geminiviruses for biotechnology: the art of parasite taming. New Phytol. 210, 58–64 (2016).
Rybicki, E. P. & Martin, D. P. in Plant Viral Vectors. Current Topics in Microbiology and Immunology Vol. 375 (eds Palmer K. & Gleba Y.) 19-45 (Springer, 2014).
Zaidi, S. S.-A. & Mansoor, S. Viral vectors for plant genome engineering. Front. Plant Sci. 8, 2012–2017 (2017).
Saunders, K., Lucy, A. & Stanley, J. DNA forms of the geminivirus African cassava mosaic virus consistent with a rolling circle mechanism of replication. Nucleic Acids Res. 19, 2325–2330 (1991).
Jeske, H., Lütgemeier, M. & Preiß, W. DNA forms indicate rolling circle and recombination-dependent replication of Abutilon mosaic virus. EMBO J. 20, 6158–6167 (2001).
Ruhel, R. & Chakraborty, S. Multifunctional roles of geminivirus encoded replication initiator protein. VirusDisease 30, 66–73 (2019).
Koonin, E. V. & Ilyina, T. V. Geminivirus replication proteins are related to prokaryotic plasmid rolling circle DNA replication initiator proteins. J. Gen. Virol. 10, 2763–2766 (1992).
Selth, L. A., Randles, J. W. & Rezaian, M. A. Agrobacterium tumefaciens supports DNA replication of diverse geminivirus types. FEBS Lett. 516, 179–182 (2002).
Lutz, K. A., Azhagiri, A. K., Tungsuchat-Huang, T. & Maliga, P. A guide to choosing vectors for transformation of the plastid genome of higher plants. Plant Physiol. 145, 1201–1210 (2007).
Stenger, D. C., Revington, G. N., Stevenson, M. C. & Bisaro, D. M. Replicational release of geminivirus genomes from tandemly repeated copies: evidence for rolling-circle replication of a plant viral DNA. Proc. Natl Acad. Sci. USA 88, 8029–8033 (1991).
Bisaro, D. M. in DNA Replication in Eukaryotic Cells (ed. DePamphilis, M. L.) Ch. 30 (Cold Spring Harbor Laboratory Press, 1996).
Svab, Z. & Maliga, P. Mutation proximal to the tRNA binding region of the Nicotiana plastid 16S rRNA confers resistance to spectinomycin. Mol. Gen. Genet. 228, 316–319 (1991).
Verma, D. & Daniell, H. Chloroplast vector systems for biotechnology applications. Plant Physiol. 145, 1129–1143 (2007).
De Cosa, B., Moar, W., Lee, S. B., Miller, M. & Daniell, H. Overexpression of the Bt cry2Aa2 operon in chloroplasts leads to formation of insecticidal crystals. Nat. Biotechnol. 19, 71–74 (2001).
Stern, D. B. & Gruissem, W. Control of plastid gene expression: 3′ inverted repeats act as mRNA processing and stabilizing elements, but do not terminate transcription. Cell 51, 1145–1157 (1987).
Shi, C. et al. Full transcription of the chloroplast genome in photosynthetic eukaryotes. Sci. Rep. 6, 30135 (2016).
Sharwood, R. E., Hotto, A. M., Bollenbach, T. J. & Stern, D. B. Overaccumulation of the chloroplast antisense RNA AS5 is correlated with decreased abundance of 5S rRNA in vivo and inefficient 5S rRNA maturation in vitro. RNA 17, 230–243 (2011).
Svab, Z. & Maliga, P. High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc. Natl Acad. Sci. USA 90, 913–917 (1993).
Tangphatsornruang, S., Birch-Machin, I., Newell, C. A. & Gray, J. C. The effect of different 3′ untranslated regions on the accumulation and stability of transcripts of a gfp transgene in chloroplasts of transplastomic tobacco. Plant Mol. Biol. 76, 385–396 (2011).
Yanisch-Perron, C., Vieira, J. & Messing, J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mpl8 and pUC19 vectors. Gene 33, 103–119 (1985).
Alting-Mees, M. A. & Short, J. M. Pbluescript II: gene mapping vectors. Nucleic Acids Res. 17, 9494 (1989).
Zhang, J. et al. Identification of cis-elements conferring high levels of gene expression in non-green plastids. Plant J. 72, 115–128 (2012).
Carrer, H., Hockenberry, T. N., Svab, Z. & Maliga, P. Kanamycin resistance as a selectable marker for plastid transformation in tobacco. Mol. Gen. Genet. 241, 49–56 (1993).
Hellens, R. P., Anne Edwards, E., Leyland, N. R., Bean, S. & Mullineaux, P. M. pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42, 819–832 (2000).
A simple and general method for transferring genes into plants. Science 227, 1229–1231 (1985).
Allen, G. C., Flores-Vergara, M. A., Krasynanski, S., Kumar, S. & Thompson, W. F. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 1, 2320–2325 (2006).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Acknowledgements
We thank X. Baudin from the ImagoSeine facility (member of the France BioImaging infrastructure supported by the French National Research Agency ANR-10-INSB-04) for his help with confocal microscopy. We thank J. P. Morris and P. Grillot for help with plant care in the greenhouse facilities. We thank H. Rothnie for proofreading our manuscript.
Author information
Authors and Affiliations
Contributions
A.J., A.S. and S.C. performed the experiments. A.J., F.V., I.M. and A.P.S. designed and analysed the experiments. A.J., I.M. and A.P.S. prepared the manuscript.
Corresponding authors
Ethics declarations
Competing interests
A.J., A.S., S.C., F.V., I.M. and A.P.S are or were employees of Algentech SAS. A.J., A.P.S. and I.M. are the authors of the patent, which was filed on the basis of this study (patent no. EP2017732382 pending).
Additional information
Peer review information Nature Plants thanks Spencer Whitney and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Phenotype of AJTB and AJTE primary transformants.
Pictures were taken 1 and 11 days post-transfer on rooting medium supplemented with transgene-selective spectinomycin. Scale bar: 1 cm.
Extended Data Fig. 2 Analysis of plants transformed by AJTB and AJTE vectors.
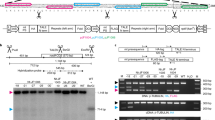
a, Physical maps of AJTB and AJTE transgene insertions in ptDNA. Localization of aadA probe, BamHI site and the resulting fragment sizes expected in the Southern blot analyses are indicated. Genes above the line are transcribed from the left to the right. Genes below the line are transcribed from the right to the left. See legend of Table 1 for construct details. b, Southern blot analysis of AJTB and AJTE lines. DNA isolated from independent lines (delimited by horizontal lines) was digested with BamHI (+) or incubated in the digestion buffer without a restriction enzyme (–) and analysed by Southern blot using aadA probe. Same DNA samples and amounts were used as in blots shown in Fig. 1. Picture of ethidium bromide-stained gel taken before transfer is shown as a loading control (total DNA). c, d, PCR analyses of AJTB and AJTE lines. Indicated pairs of primers were used for PCR analysis of the same DNA samples as in (b). The primer localizations and the expected sizes are presented in (c). The primer sequences are shown in Supplementary Table 2. The amplification products were visualized on ethidium bromide-stained agarose gels (d). No DNA (-) and WT controls were analysed in parallel.
Extended Data Fig. 3 Analyses of plants transformed by TGMV vectors.
a, Physical maps of AJTD and AJTG transgene insertions in ptDNA. Localization of transgene-specific aadA probe and plastid-specific accD and rbcL probes, relevant restriction sites and the resulting fragment sizes expected in the Southern blot analyses are indicated. Genes above the line are transcribed from the left to the right. Genes below the line are transcribed from the right to the left. Left and right targeting regions in the ptDNA are represented by horizontal double arrows. See legend of Table 1 for construct details. b–e, Southern blot analyses of AJTD and AJTG lines. DNA isolated from independent in vitro-grown lines and from WT control was digested with BamHI (+) or incubated without a restriction enzyme (–) (b), Spe1 (c) and SpeI and PvuII (d, e) and analysed by Southern blot using aadA probe (b, c), accD probe (d) or rbcL probe (e). The same DNA samples and amounts were used in all blots. Pictures of ethidium bromide-stained gels taken before transfer are shown as additional loading controls (total DNA) for panels (b) and (c). AJTG DNA highlighted by an asterisk was extracted from a green primary transformant, whereas the other AJTG samples were obtained from white plantlets.
Extended Data Fig. 4 Analyses of homoplasmic controls (HC1, HC2, HC3) and ptRep parental line.
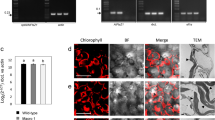
a, Physical maps of HC1, HC2, HC3 and ptRep insertions in ptDNA rbcL accD locus. Localization of transgene-specific aadA and gfp probes and plastid-specific accD and rbcL probes, relevant restriction sites and the resulting fragment sizes expected in the Southern blot analyses (and corresponding Figure numbers) are indicated. Genes above the line are transcribed from left to right. Genes below the line are transcribed from right to left. Left and right targeting regions in the ptDNA are represented by black horizontal double arrows for HC1, HC2 and ptRep insertions and by grey horizontal arrows for HC3 insertion. See legend of Table 1 for construct details. EDFig: Extended Data Fig. b, Physical map of ptDNA trnI trnA locus. Localization of plastid-specific trnI probe, relevant restriction sites and the resulting fragment sizes expected in the Southern blot analyses (and corresponding Figure numbers where trnI probe was used as a loading control) are indicated. c, DNA from WT control and from transplastomic HC1 and HC2 lines was digested with EcoRI and HindIII and analysed by Southern blot using an accD probe. d, DNA from WT control and from transplastomic ptRep and HC3 lines was digested with EcoRI and analysed by Southern blot using rbcL probe. e, Protein analysis of ptRep plants. Total proteins were extracted from a pool of in vitro-grown 3-week-old 1st (T1) or 2nd (T2) generation ptRep progeny plantlets and from WT plantlets (n≥10). Total protein staining revealed a decrease in accumulation of at least 2 proteins (indicated by asterisks), consistent with molecular weights of Rubisco large and small subunits.
Extended Data Fig. 5 Phenotype of nRep AJWY and AIBW primary transformants and transgene transmission to progeny.
a, Pictures were taken 2 weeks (nRep AJWY) or 4 weeks (AIBW) post-transfer on rooting medium supplemented with AJWY and AIBW transgene-selective spectinomycin. b, Pictures of 4-week-old nRep AJWY progeny plantlets and 8-week-old AIBW progeny plantlets grown on medium supplemented with spectinomycin (spec). Scale bar: 1 cm.
Extended Data Fig. 6 Maternal transmission of AIDR minichromosomes to the 3rd generation.
a, Physical map of the AIDR vector. Localization of the gfp probe, EcoRV site position and the resulting minichromosome size are indicated. See legend of Table 1 for construct details. b, c, Maternal transmission of minichromosomes to the 3rd generation in the absence of selective pressure. Seeds recovered from ptRep AIDR progeny plants fertilized with WT pollen were sown on a non-selective medium (lanes A) or on AIDR-selective medium (lanes B). Tobacco WT and ptRep parental line samples were analysed as controls. DNA extracted from a pool of 3-week-old plantlets (n≥10) and from a homoplasmic control (HC3) obtained by the classical insertional method (Extended Data Fig. 4a,d) was digested with EcoRI and EcoRV and analysed by Southern blot using gfp probe. The expected sizes were 2.2 kb for AIDR minichromosome and 2.5 kb for HC3. trnI-specific blot prepared in parallel using the same digestion (Extended Data Fig. 4b) is shown as a loading control (LC). The same DNA samples digested by EcoRV were analysed on an ethidium bromide-stained agarose gel (c). A band consistent with the minichromosome size is indicated by an arrow.
Extended Data Fig. 7 Maternal inheritance of kanamycin resistance for ptRep AIDR lines.
Seeds recovered from reciprocal crosses of ptRep AIDR #2 and #6 first-generation progeny plants with WT were sown on AIDR-selective medium (supplemented with kanamycin). Pictures were taken at 4 weeks post-sowing. Scale bar: 1 cm.
Extended Data Fig. 8 Minichromosome enrichment in plastidial versus total DNA isolated from a ptRep AIDR plant.
a, Physical map of the AIDR vector. Localization of the gfp probe, EcoRV site position and the resulting minichromosome size are indicated. See legend of Table 1 for construct details. b,c, Minichromosome enrichment in plastidial versus total DNA preparations. Total DNA and chloroplast enriched DNA (CP DNA) were prepared from a ptRep AIDR #6 plant. DNA was quantitated, digested with EcoRV and BamHI and subjected to serial 3-fold dilutions. The serially diluted digestions were analysed by Southern blot using a nuclear 25S rDNA probe (b, expected size 1.1kb). The same blot was subsequently hybridized with plastidial trnI probe (b, expected size 3.1kb, Extended Data Fig. 4b) and minichrosome-specific gfp probe (b, expected minichromosome size 2.2 kb). Picture of the ethidium bromide-stained gel used for Southern analysis was taken at the end of electrophoresis before transfer (c). A band consistent with the minichromosome size is indicated by an arrow.
Extended Data Fig. 9 Analyses of AIIS transplastomic lines.
a, Physical map of AIIS transgene insertion in ptDNA trnI trnA locus. Localization of transgene-specific gfp and plastid-specific trnI probes, relevant restriction sites and the resulting fragment sizes expected in the Southern blot analyses are indicated. Left and right targeting regions in the ptDNA are represented by horizontal double arrows. Genes are transcribed from left to right. Dashed arrows show expected transcripts initiated by transgene Prrn promoter (1.8 kb) and plastidial Prrn promoter (4.6 kb). See legend of Table 1 for construct details. b-d, Molecular analysis of AIIS transplastomic lines. Seeds recovered from independent AIIS transplastomic lines (line number indicated above each lane) were sown on transgene-selective medium and WT seeds were germinated on non-selective medium. b, DNA isolated from a pool of 2-week-old seedlings (n≥10) and from a non-transformed plant (WT) was digested with EcoRV and HindIII and analysed by Southern blot using trnI probe. c, RNA was extracted from a pool of 3-week-old plantlets (n≥10) and analysed by northern blot using gfp probe. The arrows indicate bands consistent with the expected transcript sizes produced from the transgene promoter (1.8 kb) and from the plastidial upstream Prrn promoter (4.8 kb) (Extended Data Fig. 9a). The lower panel shows the same blot stained with methylene blue to illustrate the abundance of rRNA as a loading control. d, Total soluble proteins were extracted from a pool of 3-week-old seedlings (n≥10) and subjected to western blotting using anti-GFP antibody (d, upper panel). After detection, the blot was stained with Coomassie dye and is shown as a loading control (d, lower panel). A band consistent with the expected GFP molecular weight (27 kDa) visible on Coomassie stained blot is highlighted by an arrow. Line numbers are indicated above the blot.
Extended Data Fig. 10 Analyses of GFP expression in individual ptRep AIDR and AIIS progeny plants.
Seeds recovered from independent ptRep AIDR minichromosome lines and AIIS insertion lines were sown on a transgene-selective medium and WT seeds were germinated on a non-selective medium and plantlets were transferred to greenhouse 2 weeks post-sowing. a, RNA extracted from youngest leaves of individual 7-week-old plants was analysed by northern blot using a gfp probe. The arrows indicate bands consistent with the expected transcript sizes produced from the transgene promoter (1.8 kb) and from plastidial upstream Prrn promoter (4.8 kb) for AIIS plants (Extended Data Fig. 9a). The lower panel shows the same blot stained with methylene blue to illustrate the abundance of rRNA as a loading control. DNA contamination of RNA samples was measured using Qubit DNA BR reagents and ranged from 20 to 40 ng in the shown ptRep AIDR RNA samples. 50ng of total DNA extracted from the same 7-week-old ptRep AIDR#K6 plant was analysed in parallel and a faint band (shown by an asterisk) was detected, indicating a minor DNA contribution to RNA specific signal. b, Total soluble proteins were extracted in parallel from the same leaves as used for RNA isolation. 6 μg and 3 μg of total proteins were subjected to western blotting using anti-GFP antibody (b, upper panel). Serially diluted GFP standard was used to quantitate GFP amounts in plant extracts. After detection, the blot was stained with Coomassie dye and is shown as a loading control (b, lower panel). Line numbers are indicated above the blots.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2 and Methods.
Source data
Source Data Fig. 1
Unprocessed Southern blots and total DNA staining.
Source Data Fig. 2
Unprocessed Southern blots.
Source Data Fig. 3
Unprocessed Southern blots.
Source Data Fig. 5
Unprocessed Southern blots and total DNA staining.
Source Data Fig. 7
Unprocessed northern blots, total RNA staining, unprocessed western blots and total protein staining.
Source Data Extended Data Fig. 2
Unprocessed Southern blots and total DNA staining.
Source Data Extended Data Fig. 3
Unprocessed Southern blots and total DNA staining.
Source Data Extended Data Fig. 4
Unprocessed Southern blots and unprocessed total protein gel.
Source Data Extended Data Fig. 6
Unprocessed Southern blots and total DNA staining.
Source Data Extended Data Fig. 8
Unprocessed Southern blots and total DNA staining.
Source Data Extended Data Fig. 9
Unprocessed Southern blot, unprocessed northern blot, total RNA staining, unprocessed western blot and total protein staining.
Source Data Extended Data Fig. 10
Unprocessed northern blot, total RNA staining, unprocessed western blot and total protein staining.
Rights and permissions
About this article
Cite this article
Jakubiec, A., Sarokina, A., Choinard, S. et al. Replicating minichromosomes as a new tool for plastid genome engineering. Nat. Plants 7, 932–941 (2021). https://doi.org/10.1038/s41477-021-00940-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-021-00940-y
This article is cited by
-
Plastid engineering using episomal DNA
Plant Cell Reports (2023)
-
Engineering the plastid and mitochondrial genomes of flowering plants
Nature Plants (2022)
-
Advances in plastid transformation for metabolic engineering in higher plants
aBIOTECH (2022)