Abstract
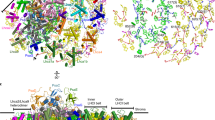
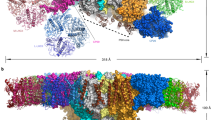
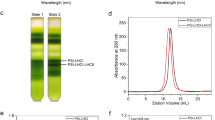
During oxygenic photosynthesis, photosystems I and II (PSI and PSII) are essential for light-driven electron transport. Excitation energy transfer in PSI occurs extremely quickly, making it an efficient energy converter. In the alga Chlamydomonas reinhardtii (Cr), multiple units of light-harvesting complex I (LHCI) bind to the PSI core and function as peripheral antennae, forming a PSI–LHCI supercomplex. CrPSI–LHCI shows significantly larger antennae compared with plant PSI–LHCI while maintaining highly efficient energy transfer from LHCI to PSI. Here, we report structures of CrPSI–LHCI, solved by cryo-electron microscopy, revealing that up to ten LHCIs are associated with the PSI core. The structures provide detailed information about antenna organization and pigment arrangement within the supercomplexes. Highly populated and closely associated chlorophylls in the antennae explain the high efficiency of light harvesting and excitation energy transfer in CrPSI–LHCI.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The atomic coordinates of the PSI–LHCI supercomplexes have been deposited in the PDB under accession codes 6IJJ (for PSI–8LHCI) and 6IJO (for PSI–10LHCI). The cryo-EM maps of these supercomplexes have been deposited in the EMDB with accession codes EMD-9678 (for PSI–8LHCI) and EMD-9680 (for PSI–10LHCI), respectively. All other data generated or analysed are available from the corresponding authors on reasonable request.
References
Nelson, N. & Ben-Shem, A. The complex architecture of oxygenic photosynthesis. Nat. Rev. Mol. Cell Biol. 5, 971–982 (2004).
Dekker, J. P. & Boekema, E. J. Supramolecular organization of thylakoid membrane proteins in green plants. Biochim. Biophys. Acta 1706, 12–39 (2005).
Nelson, N. & Junge, W. Structure and energy transfer in photosystems of oxygenic photosynthesis. Annu. Rev. Biochem. 84, 659–683 (2015).
Nelson, N. & Yocum, C. F. Structure and function of photosystems I and II. Annu. Rev. Plant. Biol. 57, 521–565 (2006).
Busch, A. & Hippler, M. The structure and function of eukaryotic photosystem I. Biochim. Biophys. Acta 1807, 864–877 (2011).
Neilson, J. A. & Durnford, D. G. Structural and functional diversification of the light-harvesting complexes in photosynthetic eukaryotes. Photosynth. Res. 106, 57–71 (2010).
Le Quiniou, C. et al. PSI-LHCI of Chlamydomonas reinhardtii: increasing the absorption cross section without losing efficiency. Biochim. Biophys. Acta 1847, 458–467 (2015).
Nelson, N. Plant photosystem I—the most efficient nano-photochemical machine. J. Nanosci. Nanotechnol. 9, 1709–1713 (2009).
Pi, X. et al. Unique organization of photosystem I-light-harvesting supercomplex revealed by cryo-EM from a red alga. Proc. Natl Acad. Sci. USA 115, 4423–4428 (2018).
Antoshvili, M., Caspy, I., Hippler, M. & Nelson, N. Structure and function of photosystem I in Cyanidioschyzon merolae. Photosynth. Res. 139, 499–508 (2018).
Mazor, Y., Borovikova, A., Caspy, I. & Nelson, N. Structure of the plant photosystem I supercomplex at 2.6 A resolution. Nat. Plants 3, 17014 (2017).
Qin, X., Suga, M., Kuang, T. & Shen, J. R. Structural basis for energy transfer pathways in the plant PSI-LHCI supercomplex. Science 348, 989–995 (2015).
Mozzo, M. et al. Functional analysis of photosystem I light-harvesting complexes (Lhca) gene products of Chlamydomonas reinhardtii. Biochim. Biophys. Acta 1797, 212–221 (2010).
Jensen, P. E., Haldrup, A., Rosgaard, L. & Scheller, H. V. Molecular dissection of photosystem I in higher plants: topology, structure and function. Physiol. Plant. 119, 313–321 (2003).
Drop, B. et al. Photosystem I of Chlamydomonas reinhardtii contains nine light-harvesting complexes (Lhca) located on one side of the core. J. Biol. Chem. 286, 44878–44887 (2011).
Subramanyam, R., Jolley, C., Brune, D. C., Fromme, P. & Webber, A. N. Characterization of a novel photosystem I-LHCI supercomplex isolated from Chlamydomonas reinhardtii under anaerobic (State II) conditions. FEBS Lett. 580, 233–238 (2006).
Kargul, J., Nield, J. & Barber, J. Three-dimensional reconstruction of a light-harvesting complex I-photosystem I (LHCI-PSI) supercomplex from the green alga Chlamydomonas reinhardtii. Insights into light harvesting for PSI. J. Biol. Chem. 278, 16135–16141 (2003).
Germano, M. et al. Supramolecular organization of photosystem I and light-harvesting complex I in Chlamydomonas reinhardtii. FEBS Lett. 525, 121–125 (2002).
Elrad, D. & Grossman, A. R. A genome’s-eye view of the light-harvesting polypeptides of Chlamydomonas reinhardtii. Curr. Genet. 45, 61–75 (2004).
Stauber, E. J. et al. Proteomics of Chlamydomonas reinhardtii light-harvesting proteins. Eukaryot. Cell 2, 978–994 (2003).
Stauber, E. J., Busch, A., Naumann, B., Svatos, A. & Hippler, M. Proteotypic profiling of LHCI from Chlamydomonas reinhardtii provides new insights into structure and function of the complex. Proteomics 9, 398–408 (2009).
Yadavalli, V., Malleda, C. & Subramanyam, R. Protein-protein interactions by molecular modeling and biochemical characterization of PSI-LHCI supercomplexes from Chlamydomonas reinhardtii. Mol. Biosyst. 7, 3143–3151 (2011).
Ozawa, S. et al. Configuration of ten light-harvesting chlorophyll a/b complex I subunits in Chlamydomonas reinhardtii photosystem I. Plant Physiol. 178, 583–595 (2018).
Pan, X. et al. Structure of the maize photosystem I supercomplex with light-harvesting complexes I and II. Science 360, 1109–1113 (2018).
Malavath, T., Caspy, I., Netzer-El, S. Y., Klaiman, D. & Nelson, N. Structure and function of wild-type and subunit-depleted photosystem I in Synechocystis. Biochim. Biophys. Acta Bioenerg. 1859, 645–654 (2018).
Caspy, I. & Nelson, N. Structure of the plant photosystem I. Biochem. Soc. Trans. 46, 285–294 (2018).
Jordan, P. et al. Three-dimensional structure of cyanobacterial photosystem I at 2.5 A resolution. Nature 411, 909–917 (2001).
Vanselow, C., Weber, A. P., Krause, K. & Fromme, P. Genetic analysis of the photosystem I subunits from the red alga, Galdieria sulphuraria. Biochim. Biophys. Acta 1787, 46–59 (2009).
Takahashi, Y., Yasui, T. A., Stauber, E. J. & Hippler, M. Comparison of the subunit compositions of the PSI-LHCI supercomplex and the LHCI in the green alga Chlamydomonas reinhardtii. Biochemistry 43, 7816–7823 (2004).
Liu, Z. et al. Crystal structure of spinach major light-harvesting complex at 2.72 A resolution. Nature 428, 287–292 (2004).
Bonente, G., Pippa, S., Castellano, S., Bassi, R. & Ballottari, M. Acclimation of Chlamydomonas reinhardtii to different growth irradiances. J. Biol. Chem. 287, 5833–5847 (2012).
Niyogi, K. K., Bjorkman, O. & Grossman, A. R. The roles of specific xanthophylls in photoprotection. Proc. Natl Acad. Sci. USA 94, 14162–14167 (1997).
Pascal, A. A. et al. Molecular basis of photoprotection and control of photosynthetic light-harvesting. Nature 436, 134–137 (2005).
Morosinotto, T., Breton, J., Bassi, R. & Croce, R. The nature of a chlorophyll ligand in Lhca proteins determines the far red fluorescence emission typical of photosystem I. J. Biol. Chem. 278, 49223–49229 (2003).
Steinbeck, J. et al. Structure of a PSI-LHCI-cyt b 6 f supercomplex in Chlamydomonas reinhardtii promoting cyclic electron flow under anaerobic conditions. Proc. Natl Acad. Sci. USA 115, 10517–10522 (2018).
Gorman, D. S. & Levine, R. P. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc. Natl Acad. Sci. USA 54, 1665–1669 (1965).
Fischer, N., Setif, P. & Rochaix, J. D. Targeted mutations in the psaC gene of Chlamydomonas reinhardtii: preferential reduction of FB at low temperature is not accompanied by altered electron flow from photosystem I to ferredoxin. Biochemistry 36, 93–102 (1997).
Chua, N. H. & Bennoun, P. Thylakoid membrane polypeptides of Chlamydomonas reinhardtii: wild-type and mutant strains deficient in photosystem II reaction center. Proc. Natl Acad. Sci. USA 72, 2175–2179 (1975).
Wei, X. et al. Structure of spinach photosystem II-LHCII supercomplex at 3.2 A resolution. Nature 534, 69–74 (2016).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Zhang, J. et al. Structure of phycobilisome from the red alga Griffithsia pacifica. Nature 551, 57–63 (2017).
Grant, T. & Grigorieff, N. Automatic estimation and correction of anisotropic magnification distortion in electron microscopes. J. Struct. Biol. 192, 204–208 (2015).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Kimanius, D., Forsberg, B. O., Scheres, S.H. & Lindahl, E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. eLife 5, e18722 (2016).
Zhu, D. et al. Pushing the resolution limit by correcting the Ewald sphere effect in single-particle cryo-EM reconstructions. Nat. Commun. 9, 1552 (2018).
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D67, 235–242 (2011).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D66, 213–221 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D66, 12–21 (2010).
Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014).
Acknowledgements
Cryo-EM data collection was carried out at the Center for Biological Imaging, Core Facilities for Protein Science at the Institute of Biophysics, Chinese Academy of Sciences. We thank W. Yang from the Institute of Botany, Chinese Academy of Sciences, for providing the C. reinhardtii CC-124 strain; B. Zhu, X. Huang, G. Ji, D. Fan, T. Niu, F. Sun and other staff members at the Center for Biological Imaging for their support in data collection; L. Niu, X. Ding, M. Zhang and F. Yang for mass spectrometry; and L. Kong for cryo-EM data storage and backup. The project was funded by the National Key R&D Program of China (nos. 2017YFA0504700, 2017YFA0503702 and 2016YFA0502900), the Strategic Priority Research Program at the Chinese Academy of Sciences (nos. XDB08020302, XDB08030204 and XDB27020106), the Key Research Program of Frontier Sciences at the Chinese Academy of Sciences (no. QYZDB-SSW-SMC005) and the National Natural Science Foundation of China (nos. 31770778, 31700649 and 31600609). Z.L. and X. Zhang received scholarships from the ‘National Thousand Young Talents Program’ from the Office of Global Experts Recruitment in China. X.S., X.P. and J.M. were sponsored by the Youth Innovation Promotion Association at the Chinese Academy of Sciences.
Author information
Authors and Affiliations
Contributions
M.L., W.C. and X.Zhang conceived the project. X.Zhao cultured the algae. X.S. performed sample preparation and characterization. X.S. and X.P. collected the cryo-EM data. J.M. and X.Zhang processed the cryo-EM data and reconstructed the cryo-EM maps. X.P. built and refined the structure model. X.P., M.L., X.Zhang and Z.L. analysed the structure. M.L., X.Zhang and Z.L. wrote the manuscript. All authors discussed and commented on the results and the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Journal peer review information: Nature Plants thanks Egbert Boekema and Jean-David Rochaix and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–8 and Supplementary Tables 1–3.
Rights and permissions
About this article
Cite this article
Su, X., Ma, J., Pan, X. et al. Antenna arrangement and energy transfer pathways of a green algal photosystem-I–LHCI supercomplex. Nat. Plants 5, 273–281 (2019). https://doi.org/10.1038/s41477-019-0380-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-019-0380-5
This article is cited by
-
Uncovering the photosystem I assembly pathway in land plants
Nature Plants (2024)
-
Architecture of symbiotic dinoflagellate photosystem I–light-harvesting supercomplex in Symbiodinium
Nature Communications (2024)
-
Structural insights into a unique PSI–LHCI–LHCII–Lhcb9 supercomplex from moss Physcomitrium patens
Nature Plants (2023)
-
Structural insights into the assembly and energy transfer of the Lhcb9-dependent photosystem I from moss Physcomitrium patens
Nature Plants (2023)
-
Structural insights into photosystem II supercomplex and trimeric FCP antennae of a centric diatom Cyclotella meneghiniana
Nature Communications (2023)