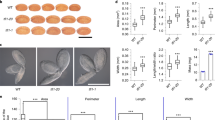
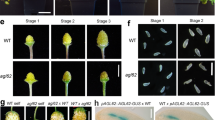
Abstract
The angiosperm seed is composed of three genetically distinct tissues: the diploid embryo that originates from the fertilized egg cell, the triploid endosperm that is produced from the fertilized central cell, and the maternal sporophytic integuments that develop into the seed coat1. At the onset of embryo development in Arabidopsis thaliana, the zygote divides asymmetrically, producing a small apical embryonic cell and a larger basal cell that connects the embryo to the maternal tissue2. The coordinated and synchronous development of the embryo and the surrounding integuments, and the alignment of their growth axes, suggest communication between maternal tissues and the embryo. In contrast to animals, however, where a network of maternal factors that direct embryo patterning have been identified3,4, only a few maternal mutations have been described to affect embryo development in plants5,6,7. Early embryo patterning in Arabidopsis requires accumulation of the phytohormone auxin in the apical cell by directed transport from the suspensor8,9,10. However, the origin of this auxin has remained obscure. Here we investigate the source of auxin for early embryogenesis and provide evidence that the mother plant coordinates seed development by supplying auxin to the early embryo from the integuments of the ovule. We show that auxin response increases in ovules after fertilization, due to upregulated auxin biosynthesis in the integuments, and this maternally produced auxin is required for correct embryo development.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Figueiredo, D. D. & Köhler, C. C. Auxin: a molecular trigger of seed development. Genes Dev. 32, 479–490 (2018).
Mansfield, S. G. & Briarty, L. G. Early embryogenesis in Arabidopsis thaliana. 2. The developing embryo. Can. J. Bot.-Rev. Can. De. Bot. 69, 461–476 (1991).
Johnstone, O. & Lasko, P. Translational regulation and RNA localization in Drosophila oocytes and embryos. Annu. Rev. Genet. 35, 365–406 (2001).
Riechmann, V. & Ephrussi, A. Axis formation during Drosophila oogenesis. Curr. Opin. Gen. Dev. 11, 374–383 (2001).
Ray, S., Golden, T. & Ray, A. Maternal effects of the short integument mutation on embryo development in. Arab. Dev. Biol. 180, 365–369 (1995).
Prigge, M. J. & Wagner, D. R. The Arabidopsis SERRATE gene encodes a zinc-finger protein required for normal shoot development. Plant Cell 13, 1263–1280 (2001).
Costa, L. M. et al. Central cell-derived peptides regulate early embryo patterning in flowering plants. Science 344, 168–172 (2014).
Möller, B. K. & Weijers, D. Auxin control of embryo patterning. Cold Spring Harb. Perspect. Biol. 1, a001545 (2009).
Friml, J. et al. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426, 147–153 (2003).
Robert, H. S. et al. Local auxin sources orient the apical-basal axis in Arabidopsis embryos. Curr. Biol. 23, 2506–2512 (2013).
Brunoud, G. et al. A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482, 103–106 (2012).
Liao, C.-Y. et al. Reporters for sensitive and quantitative measurement of auxin response. Nat. Methods 12, 207–210 (2015).
Paciorek, T. & Friml, J. Auxin signaling. J. Cell Sci. 119, 1199–1202 (2006).
Ljung, K. Auxin metabolism and homeostasis during plant development. Development 140, 943–950 (2013).
Stepanova, A. N. et al. The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 23, 3961–3973 (2011).
Mashiguchi, K. et al. The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl Acad. Sci. USA 108, 18512–18517 (2011).
Won, C. et al. Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc. Natl Acad. Sci. USA 108, 18518–18523 (2011).
Stepanova, A. N. et al. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133, 177–191 (2008).
Jensen, P. J., Hangarter, R. P. & Estelle, M. Auxin transport is required for hypocotyl elongation in light-grown but not dark-grown Arabidopsis. Plant Physiol. 116, 455–462 (1998).
Debeaujon, I. et al. Proanthocyanidin-accumulating cells in Arabidopsis testa: regulation of differentiation and role in seed development. Plant Cell 15, 2514–2531 (2003).
Figueiredo, D. D., Batista, R. A., Roszak, P. J., & Köhler, C. C. Auxin production couples endosperm development to fertilization. Nat. Plants 1, 15184 (2015).
Figueiredo, D. D., Batista, R. A., Roszak, P. J., Hennig, L. & Köhler, C. C. Auxin production in the endosperm drives seed coat development in Arabidopsis. eLife 5, e20542 (2016).
Blilou, I. et al. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433, 39–44 (2005).
Vieten, A. et al. Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 132, 4521–4531 (2005).
Weijers, D. et al. Auxin triggers transient local signaling for cell specification in Arabidopsis embryogenesis. Dev. Cell 10, 265–270 (2006).
Larsson, E., Vivian-Smith, A., Offringa, R. & Sundberg, E. Auxin homeostasis in Arabidopsis ovules is anther-dependent at maturation and changes dynamically upon fertilization. Front. Plant Sci. 8, 315–14 (2017).
Weijers, D. et al. Maintenance of embryonic auxin distribution for apical-basal patterning by PIN-FORMED-dependent auxin transport in Arabidopsis. Plant Cell 17, 2517–2526 (2005).
Wabnik, K., Robert, H. S., Smith, R. S. & Friml, J. Modeling framework for the establishment of the apical-basal embryonic axis in plants. Curr. Biol. 23, 2513–2518 (2013).
Prat, T. et al. WRKY23 is a component of the transcriptional network mediating auxin feedback on PIN polarity. PLoS Genet. 14, e1007177 (2018).
Gallavotti, A., Yang, Y., Schmidt, R. J. & Jackson, D. P. The relationship between auxin transport and maize branching. Plant Physiol. 147, 1913–1923 (2008).
Zhang, J., & Peer, W. A. Auxin homeostasis: the DAO of catabolism. J. Exp. Bot. 68, 3145–3154 (2017).
Pencík, A. et al. Ultra-rapid auxin metabolite profiling for high-throughput mutant screening in Arabidopsis. J. Exp. Bot. 69, 2569–2579 (2018).
Acknowledgements
We thank E. Groot for assistance with statistical analyses and for critical reading of the manuscript. We thank J. Alonso for providing wei8-1, wei8-1 tar1-1, pDR5:GFP wei8-1, and pTAA1:GFP-TAA1 seeds, G. Jürgens for providing pDR5:nls-3xGFP and pDR5:nls-3xGFP wei8-1 tar1-1 seeds, T. Vernoux for p35S:DII-VENUS and p35S:mDII-VENUS seeds and plasmids, R. Offringa for pSDM7010 and pSDM7012, L. Lepiniec for pBAN-pBS-SK, C.-Y. Liao and D. Weijers for R2D2 seeds, and T. Friedrich for valuable suggestions. Seeds of wei8-3 and wei-11 seeds were obtained from the European Arabidopsis Stock Center (NASC). We acknowledge the CEITEC core facility CELLIM supported by the MEYS CR (LM2015062 Czech-BioImaging), and the CEITEC core facility Plant Sciences. H.S.R. was supported by the SoMoProII program co-financed by the South-Moravian Region and the European Union, by the Ministry of Education, Youth and Sports of the Czech Republic within CEITEC 2020 (LQ1601) and by the Masaryk University. C.P. was supported by the Kwanjeong Educational Foundation. C.L.G. was supported by the Deutscher Akademischer Austauschdienst. W.G. was a post-doctoral fellow of the Research Foundation Flanders. B.W. was supported by the ‘NITKA’ project under European Social Fund UDA-POKL.04.03.00-00-168/12, realized at the University of Silesia, Katowice, Poland. A.P and O.N. were supported by the Czech Foundation Agency (GA17-21581Y) and the Ministry of Education, Youth and Sports of the Czech Republic via the National Program for Sustainability (LO1204). J.C. is a post-doctoral fellow supported by a grant from the Deutsche Forschungsgemeinschaft (Dr334/10) to T.D. This work was further supported by the European Research Council (FP7/2007-2013 / ERC-grant agreement no. 282300 to J.F.), and the Czech Science Foundation GACR (GA13-40637S) to J.F.; and by the Deutsche Forschungsgemeinschaft (La606/6, La606/13, La606/17 and ERA-CAPS program), and the EU 7th framework program (ITN SIREN) to T.L. This material reflects only the author’s views and the European Union is not liable for any use that may be made of the information contained therein.
Author information
Authors and Affiliations
Contributions
H.S.R., C.P. and C.L.G. contributed equally to this work and performed experiments. H.S.R., B.W., A.P. and O.N. collected samples and performed the analysis for the auxin measurements. W.G. participated in the backcrosses experiments. J.C. and T.D. provided the maize data. H.S.R., C.P., C.L.G., J.F. and T.L. designed the experiments, analysed the data and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Impact statement:
Early embryo development requires auxin production in the surrounding maternal integuments.
Supplementary information
Supplementary Information
Supplementary Materials, Supplementary Figures 1–6, Supplementary Tables 1–3 and Supplementary References
Rights and permissions
About this article
Cite this article
Robert, H.S., Park, C., Gutièrrez, C.L. et al. Maternal auxin supply contributes to early embryo patterning in Arabidopsis. Nature Plants 4, 548–553 (2018). https://doi.org/10.1038/s41477-018-0204-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-018-0204-z
This article is cited by
-
The orchid seed coat: a developmental and functional perspective
Botanical Studies (2023)
-
Carbon and nitrogen metabolism affects kentucky bluegrass rhizome expansion
BMC Plant Biology (2023)
-
Maternal control of triploid seed development by the TRANSPARENT TESTA 8 (TT8) transcription factor in Arabidopsis thaliana
Scientific Reports (2023)
-
Transcriptome analysis of ovules offers early developmental clues after fertilization in Cicer arietinum L.
3 Biotech (2023)
-
AUX1, PIN3, and TAA1 collectively maintain fertility in Arabidopsis
Planta (2023)