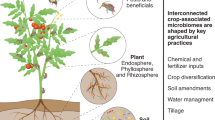
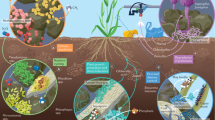
Abstract
In an era of ecosystem degradation and climate change, maximizing microbial functions in agroecosystems has become a prerequisite for the future of global agriculture. However, managing species-rich communities of plant-associated microbiomes remains a major challenge. Here, we propose interdisciplinary research strategies to optimize microbiome functions in agroecosystems. Informatics now allows us to identify members and characteristics of ‘core microbiomes’, which may be deployed to organize otherwise uncontrollable dynamics of resident microbiomes. Integration of microfluidics, robotics and machine learning provides novel ways to capitalize on core microbiomes for increasing resource-efficiency and stress-resistance of agroecosystems.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
14 August 2018
Owing to a technical error, this Perspective was originally published without its received and accepted dates; the dates “Received: 31 December 2017; Accepted: 23 March 2018” have now been included in all versions.
References
Berendsen, R. L., Pieterse, C. M. & Bakker, P. A. The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486 (2012).
de Vries, F. & Wallenstein, M. Below-ground connections underlying above-ground food production: a framework for optimising ecological connections in the rhizosphere. J. Ecol. 105, 913–920 (2017).
Busby, P. E. et al. Research priorities for harnessing plant microbiomes in sustainable agriculture. PLOS Biol. 15, e2001793 (2017).
Elser, J. & Bennett, E. Phosphorus cycle: a broken biogeochemical cycle. Nature 478, 29–31 (2011).
Callaway, E. Devastating wheat fungus appears in Asia for first time. Nature 532, 421–422 (2016).
Howden, S. M. et al. Adapting agriculture to climate change. Proc. Natl Acad. Sci. USA 104, 19691–19696 (2007).
Dangl, J. L., Horvath, D. M. & Staskawicz, B. J. Pivoting the plant immune system from dissection to deployment. Science 341, 746–751 (2013).
Robertson, G. P. & Vitousek, P. M. Nitrogen in agriculture: balancing the cost of an essential resource. Ann. Rev. Env. Res. 34, 97–125 (2009).
Tilman, D., Balzer, C., Hill, J. & Befort, B. L. Global food demand and the sustainable intensification of agriculture. Proc. Natl Acad. Sci. USA 108, 20260–20264 (2011).
van der Heijden, M. G. et al. Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature 396, 69–72 (1998).
Bonfante, P. & Anca, I.-A. Plants, mycorrhizal fungi, and bacteria: a network of interactions. Ann. Rev. Microbiol. 63, 363–383 (2009).
Hiruma, K. et al. Root endophyte Colletotrichum tofieldiae confers plant fitness benefits that are phosphate status dependent. Cell 165, 464–474 (2016).
Mendes, R. et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332, 1097–1100 (2011).
Zavala-Gonzalez, E. A. et al. Arabidopsis thaliana root colonization by the nematophagous fungus Pochonia chlamydosporia is modulated by jasmonate signaling and leads to accelerated flowering and improved yield. New Phytol. 213, 351–364 (2017).
Calvo, P., Nelson, L. & Kloepper, J. W. Agricultural uses of plant biostimulants. Plant Soil 383, 3–41 (2014).
Arnold, A. E. et al. Fungal endophytes limit pathogen damage in a tropical tree. Proc. Natl Acad. Sci. USA 100, 15649–15654 (2003).
Vorholt, J. A., Vogel, C., Carlström, C. I. & Müller, D. B. Establishing causality: opportunities of synthetic communities for plant microbiome research. Cell Host Microbe 22, 142–155 (2017).
Robertson, G. P. et al. Cellulosic biofuel contributions to a sustainable energy future: choices and outcomes. Science 356, eaal2324 (2017).
Castrillo, G. et al. Root microbiota drive direct integration of phosphate stress and immunity. Nature 543, 513–518 (2017).
Jacott, C. N., Murray, J. D. & Ridout, C. J. Trade-offs in arbuscular mycorrhizal symbiosis: disease resistance, growth responses and perspectives for crop breeding. Agronomy 7, 75 (2017).
Oldroyd, G. E. Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 11, 252–263 (2013).
King, A. The future of agriculture. Nature 544, 21–23 (2017).
Schlaeppi, K. & Bulgarelli, D. The plant microbiome at work. Mol. Plant-Microbe Int. 28, 212–217 (2015).
Girlanda, M. et al. Impact of biocontrol Pseudomonas fluorescens CHA0 and a genetically modified derivative on the diversity of culturable fungi in the cucumber rhizosphere. Appl. Env. Micriobiol. 67, 1851–1864 (2001).
Streeter, J. G. Failure of inoculant rhizobia to overcome the dominance of indigenous strains for nodule formation. Can. J. Microbiol. 40, 513–522 (1994).
Castro-Sowinski, S., Herschkovitz, Y., Okon, Y. & Jurkevitch, E. Effects of inoculation with plant growth-promoting rhizobacteria on resident rhizosphere microorganisms. FEMS Microbiol. Lett. 276, 1–11 (2007).
Walsh, U. et al. Residual impact of the biocontrol inoculant Pseudomonas fluorescens F113 on the resident population of rhizobia nodulating a red clover rotation crop. Microb. Ecol. 45, 145–155 (2003).
Lloyd-Price, J. et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 550, 61–66 (2017).
Bashan, A. et al. Universality of human microbial dynamics. Nature 534, 259–262 (2016).
Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K. & Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230 (2012).
Paramsothy, S. et al. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. Lancet 389, 1218–1228 (2017).
Müller, D. B., Vogel, C., Bai, Y. & Vorholt, J. A. The plant microbiota: systems-level insights and perspectives. Ann., Rev. Genet. 50, 211–234 (2016).
Toju, H., Yamamoto, S., Tanabe, A. S., Hayakawa, T. & Ishii, H. S. Network modules and hubs in plant-root fungal biomes. J. R. Soc. Interface 13, 20151097 (2016).
Bulgarelli, D. et al. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe 17, 392–403 (2015).
Marasco, R., Rolli, E., Fusi, M., Michoud, G. & Daffonchio, D. Grapevine rootstocks shape underground bacterial microbiome and networking but not potential functionality. Microbiome 6, 3 (2018).
Edwards, J. et al. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl Acad. Sci. USA 112, 911–920 (2015).
Hartman, K. et al. Cropping practices manipulate abundance patterns of root and soil microbiome members paving the way to smart farming. Microbiome 6, 14 (2018).
Arumugam, M. et al. Enterotypes of the human gut microbiome. Nature 473, 174–180 (2011).
Knights, D. et al. Rethinking “enterotypes”. Cell Host Microbe 16, 433–437 (2014).
Scheffer, M., Carpenter, S., Foley, J. A., Folke, C. & Walker, B. Catastrophic shifts in ecosystems. Nature 413, 591–596 (2001).
Beisner, B. E., Haydon, D. T. & Cuddington, K. Alternative stable states in ecology. Front. Ecol. Env. 1, 376–382 (2003).
Kim, S.-W. et al. Robustness of gut microbiota of healthy adults in response to probiotic intervention revealed by high-throughput pyrosequencing. DNA Res. 20, 241–253 (2013).
Dethlefsen, L. & Relman, D. A. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl Acad. Sci. USA 108, 4554–4561 (2011).
Ng, K. M. et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 502, 96–99 (2013).
Hacquard, S. et al. Microbiota and host nutrition across plant and animal kingdoms. Cell Host Microbe 17, 603–616 (2015).
Werner, G. D. & Kiers, E. T. Order of arrival structures arbuscular mycorrhizal colonization of plants. New Phytol. 205, 1515–1524 (2015).
Braun-Kiewnick, A., Jacobsen, B. J. & Sands, D. C. Biological control of Pseudomonas syringae pv. syringae, the causal agent of basal kernel blight of barley, by antagonistic Pantoea agglomerans. Phytopathology 90, 368–375 (2000).
Fukami, T. Historical contingency in community assembly: integrating niches, species pools, and priority effects. Ann. Rev. Ecol. Evol. Syst. 46, 1–23 (2015).
Wei, Z. et al. Trophic network architecture of root-associated bacterial communities determines pathogen invasion and plant health. Nat. Commun. 6, 8413 (2015).
Weller, D. M. Biological control of soilborne plant pathogens in the rhizosphere with bacteria. Ann. Rev. Phytopathol. 26, 379–407 (1988).
Morris, C. E. & Monier, J.-M. The ecological significance of biofilm formation by plant-associated bacteria. Ann. Rev. Phytopathol. 41, 429–453 (2003).
Pieterse, C. M. et al. Induced systemic resistance by beneficial microbes. Ann. Rev. Phytopathol. 52, 347–375 (2014).
Wrather, J. A. & Koenning, S. R. Effects of diseases on soybean yields in the United States 1996 to 2007. Plant Health Prog. https://doi.org/10.1094/PHP-2009-0401-01-RS (2009).
Freilich, S. et al. Competitive and cooperative metabolic interactions in bacterial communities. Nat. Commun. 2, 589 (2011).
Masciarelli, O., Llanes, A. & Luna, V. A new PGPR co-inoculated with Bradyrhizobium japonicum enhances soybean nodulation. Microbiol. Res. 169, 609–615 (2014).
Cassan, F. et al. Azospirillum brasilense Az39 and Bradyrhizobium japonicum E109, inoculated singly or in combination, promote seed germination and early seedling growth in corn (Zea mays L.) and soybean (Glycine max L.). Eur. J. Soil Biol. 45, 28–35 (2009).
Paredes, S. H. et al. Design of synthetic bacterial communities for predictable plant phenotypes. PLOS Biol. 16, e2003962 (2018).
Lundberg, D. S. et al. Defining the core Arabidopsis thaliana root microbiome. Nature 488, 86–90 (2012).
Barea, J.-M., Pozo, M. J., Azcon, R. & Azcon-Aguilar, C. Microbial co-operation in the rhizosphere. J. Exp. Bot. 56, 1761–1778 (2005).
Agler, M. T. et al. Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLOS Biol. 14, e1002352 (2016).
Faust, K. & Raes, J. Microbial interactions: from networks to models. Nat. Rev. Microbiol. 10, 538–550 (2012).
Coyte, K. Z., Schluter, J. & Foster, K. R. The ecology of the microbiome: networks, competition, and stability. Science 350, 663–666 (2015).
Newman, M. E. J. Networks: an Introduction (Oxford University Press, New York, 2010).
Toju, H. et al. Species-rich networks and eco-evolutionary synthesis at the metacommunity level. Nat. Ecol. Evol. 1, 0024 (2017).
Bai, Y. et al. Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528, 364–369 (2015).
Layeghifard, M., Hwang, D. M. & Guttman, D. S. Disentangling interactions in the microbiome: a network perspective. Trends Microbiol. 25, 217–228 (2017).
Hartman, K., van der Heijden, M. G., Roussely-Provent, V., Walser, J.-C. & Schlaeppi, K. Deciphering composition and function of the root microbiome of a legume plant. Microbiome 5, 2 (2017).
Bianciotto, V. et al. An obligately endosymbiotic mycorrhizal fungus itself harbors obligately intracellular bacteria. Appl. Env. Micriobiol. 62, 3005–3010 (1996).
Behie, S. W. et al. Carbon translocation from a plant to an insect-pathogenic endophytic fungus. Nat. Commun. 8, 14245 (2017).
Langille, M. G. et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotech. 31, 814–821 (2013).
Callahan, B. J. et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016).
Delgado-Baquerizo, M. et al. A global atlas of the dominant bacteria found in soil. Science 359, 320–325 (2018).
Deyle, E. R., May, R. M., Munch, S. B. & Sugihara, G. Tracking and forecasting ecosystem interactions in real time. P. Roy. Soc. Lond. B Bio. 283, 20152258 (2016).
Sugihara, G. et al. Detecting causality in complex ecosystems. Science 338, 496–500 (2012).
Ushio, M. et al. Fluctuating interaction network and time-varying stability of a natural fish community. Nature 554, 360–363 (2018).
Suzuki, K., Yoshida, K., Nakanishi, Y. & Fukuda, S. An equation-free method reveals the ecological interaction networks within complex microbial ecosystems. Methods Ecol. Evol. 8, 1774–1785 (2017).
Schreiber, T. Measuring information transfer. Phys. Rev. Lett. 85, 461–464 (2000).
Chang, C.-W., Ushio, M. & Hsieh, C.-H. Empirical dynamic modeling for beginners. Ecol. Res. 32, 785–796 (2017).
Vandeputte, D. et al. Quantitative microbiome profiling links gut community variation to microbial load. Nature 551, 507–511 (2017).
Smets, W. et al. A method for simultaneous measurement of soil bacterial abundances and community composition via 16S rRNA gene sequencing. Soil Biol. Biochem. 96, 145–151 (2016).
Nguyen, N. H. et al. FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 20, 241–248 (2016).
van der Heijden, M. G., de Bruin, S., Luckerhoff, L., van Logtestijn, R. S. & Schlaeppi, K. A widespread plant-fungal-bacterial symbiosis promotes plant biodiversity, plant nutrition and seedling recruitment. ISME J. 10, 389–399 (2016).
Sheth, R. U., Cabral, V., Chen, S. P. & Wang, H. H. Manipulating bacterial communities by in situ microbiome engineering. Trends Genet. 32, 189–200 (2016).
Mee, M. T., Collins, J. J., Church, G. M. & Wang, H. H. Syntrophic exchange in synthetic microbial communities. Proc. Natl Acad. Sci. USA 111, 2149–2156 (2014).
Fondi, M. & Liò, P. Multi-omics and metabolic modelling pipelines: challenges and tools for systems microbiology. Microb. Res. 171, 52–64 (2015).
Peay, K. G. et al. Convergence and contrast in the community structure of Bacteria, Fungi and Archaea along a tropical elevation–climate gradient. FEMS Microbiol. Ecol. 93, 5 (2017).
Narisawa, K., Hambleton, S. & Currah, R. S. Heteroconium chaetospira, a dark septate root endophyte allied to the Herpotrichiellaceae (Chaetothyriales) obtained from some forest soil samples in Canada using bait plants. Mycoscience 48, 274–281 (2007).
Usuki, F. & Narisawa, K. A mutualistic symbiosis between a dark septate endophytic fungus, Heteroconium chaetospira, and a nonmycorrhizal plant, Chinese cabbage. Mycologia 99, 175–184 (2007).
Aleklett, K. et al. Build your own soil: exploring microfluidics to create microbial habitat structures. ISME J. 12, 312–319 (2018).
Li, R., Lv, X., Zhang, X., Saeed, O. & Deng, Y. Microfluidics for cell-cell interactions: A review. Front. Chem. Sci. Eng. 10, 90–98 (2016).
Nichols, D. et al. Use of ichip for high-throughput in situ cultivation of “uncultivable” microbial species. Appl. Env. Micriobiol. 76, 2445–2450 (2010).
Ikeda, S. et al. Development of a bacterial cell enrichment method and its application to the community analysis in soybean stems. Microb. Ecol. 58, 703–714 (2009).
Ikeda, S. et al. Community-and genome-based views of plant-associated bacteria: plant–bacterial interactions in soybean and rice. Plant Cell Physiol. 51, 1398–1410 (2010).
Hosokawa, M. et al. Droplet-based microfluidics for high-throughput screening of a metagenomic library for isolation of microbial enzymes. Biosens. Bioelectr. 67, 379–385 (2015).
Song, Y., Yin, H. & Huang, W. E. Raman activated cell sorting. Curr. Opin. Chem. Biol. 33, 1–8 (2016).
Grossmann, G. et al. The RootChip: an integrated microfluidic chip for plant science. Plant Cell 23, 4234–4240 (2011).
Massalha, H., Korenblum, E., Malitsky, S., Shapiro, O. H. SpringerAmpamp; Aharoni, A. Live imaging of root–bacteria interactions in a microfluidics setup. Proc. Natl Acad. Sci. USA 114, 4549–4554 (2017).
Monteagudo-Mera, A. et al. High purity galacto-oligosaccharides enhance specific Bifidobacterium species and their metabolic activity in the mouse gut microbiome. Benef. Microb. 7, 247–264 (2016).
Tilman, D., Cassman, K. G., Matson, P. A., Naylor, R. & Polasky, S. Agricultural sustainability and intensive production practices. Nature 418, 671–677 (2002).
Yachi, S. & Loreau, M. Biodiversity and ecosystem productivity in a fluctuating environment: the insurance hypothesis. Proc. Natl Acad. Sci. USA 96, 1463–1468 (1999).
Maherali, H. & Klironomos, J. N. Influence of phylogeny on fungal community assembly and ecosystem functioning. Science 316, 1746–1748 (2007).
Mundt, C. Use of multiline cultivars and cultivar mixtures for disease management. Ann. Rev. Phytopathol. 40, 381–410 (2002).
Suzuki, S. U. & Sasaki, A. How does the resistance threshold in spatially explicit epidemic dynamics depend on the basic reproductive ratio and spatial correlation of crop genotypes? J. Theor. Biol. 276, 117–125 (2011).
Isbell, F. et al. Benefits of increasing plant diversity in sustainable agroecosystems. J. Ecol. 105, 871–879 (2017).
Prieto, I. et al. Complementary effects of species and genetic diversity on productivity and stability of sown grasslands. Nat. Plants 1, 15033 (2015).
Pham, T. A. N. & Lawley, T. D. Emerging insights on intestinal dysbiosis during bacterial infections. Curr. Opin. Microbiol. 17, 67–74 (2014).
Shen, D., Wu, G. & Suk, H.-I. Deep learning in medical image analysis. Ann. Rev. Biomed. Eng. 19, 221–248 (2017).
Torkamani, A., Andersen, K. G., Steinhubl, S. R. & Topol, E. J. High-definition medicine. Cell 170, 828–843 (2017).
Scheffer, M., Carpenter, S. R., Dakos, V. & van Nes, E. H. Generic indicators of ecological resilience: Inferring the chance of a critical transition. Ann. Rev. Ecol. Evol. Syst. 46, 145–167 (2015).
Schmidt, M. & Lipson, H. Distilling free-form natural laws from experimental data. Science 324, 81–85 (2009).
Murphy, K. P. Machine Learning: a Probabilistic Perspective (MIT press, London, 2012).
Sugiura, R. et al. Field phenotyping system for the assessment of potato late blight resistance using RGB imagery from an unmanned aerial vehicle. Biosyst. Eng. 148, 1–10 (2016).
Araus, J. L. & Cairns, J. E. Field high-throughput phenotyping: the new crop breeding frontier. Trends Plant Sci. 19, 52–61 (2014).
Quesada-González, D. & Merkoçi, A. Mobile phone-based biosensing: an emerging “diagnostic and communication” technology. Biosens. Bioelectr. 92, 549–562 (2017).
Pan, S. J. & Yang, Q. A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 22, 1345–1359 (2010).
Brundrett, M. C. Coevolution of roots and mycorrhizas of land plants. New Phytol. 154, 275–304 (2002).
Foster, K. R., Schluter, J., Coyte, K. Z. & Rakoff-Nahoum, S. The evolution of the host microbiome as an ecosystem on a leash. Nature 548, 43–51 (2017).
Franzosa, E. A. et al. Relating the metatranscriptome and metagenome of the human gut. Proc. Natl Acad. Sci. USA 111, 2329–2338 (2014).
Acknowledgements
We thank Takashi Akagi and three anonymous reviewers for their insightful comments on the manuscript. This work was financially supported by JSPS KAKENHI Grant (26711026), JST PRESTO (JPMJPR16Q6), and the Funding Program for Next Generation World-Leading Researchers of Cabinet Office, the Government of Japan (GS014) to H.T, DOE Award DE-SC0016097 to KGP, and by a European Research Council Grant (335542) to E.T.K.
Author information
Authors and Affiliations
Contributions
H.T. designed the study and wrote the first draft. H.T. and E.T.K. edited the final version of the manuscript based on discussion with all the authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Toju, H., Peay, K.G., Yamamichi, M. et al. Core microbiomes for sustainable agroecosystems. Nature Plants 4, 247–257 (2018). https://doi.org/10.1038/s41477-018-0139-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-018-0139-4
This article is cited by
-
Response mechanism of soil microorganisms to simulated precipitation in the source wetland of Qinghai Lake
Ecological Processes (2024)
-
Host genotype, soil composition, and geo-climatic factors shape the fonio seed microbiome
Microbiome (2024)
-
Dynamic root microbiome sustains soybean productivity under unbalanced fertilization
Nature Communications (2024)
-
Traditional potato tillage systems in the Peruvian Andes impact bacterial diversity, evenness, community composition, and functions in soil microbiomes
Scientific Reports (2024)
-
MAPK Cascades in Plant Microbiota Structure and Functioning
Journal of Microbiology (2024)