Abstract
Rett syndrome (RTT) is an X-linked progressive and severe neurological disorder caused by mutations in the gene encoding methyl CpG binding protein 2 (MECP2). Among the 49 typical RTT patients examined, we identified 10 novel and eight known insertion/deletion variants, and 31 known pathogenic variants in MECP2. The pathogenic variants presented here should be a useful resource for examining the correlation between the genotypes and phenotypes of RTT.
Similar content being viewed by others
Rett syndrome (RTT) is a progressive neurodevelopmental disorder that affects brain development and function in females, with a prevalence of one in 10,000 worldwide1. Typical RTT is caused by mutations in the gene encoding methyl-CpG binding protein 2 (MECP2)2. A database of a large collection of MECP2 variants was established in 2002 (RettBASE: http://mecp2.chw.edu.au/index.shtml)3. To date, associations between clinical phenotypes and related genetic variants for MECP2 as well as other RTT-associated genes, including CDKL5 and FOXG1, are available.
Here, we report a total of 49 RTT patients with 10 novel insertion/deletion variants, eight known insertion/deletion variants and 31 known pathological variants.
All patients were diagnosed with typical RTT by Japanese child neurology experts according to the international diagnostic criteria for RTT. Clinical information and samples from the patient and parents were obtained with written informed consent. The study was approved by the ethical committee of NCNP. Genomic DNA was extracted from peripheral blood using a standard protocol. We first used the MLPA method (MRC-Holland, DL Amsterdam, The Netherlands) to identify the structural abnormalities in the MECP2 locus. In the patients excluded for structural abnormalities, we amplified all coding exons of MECP2 and their exon-intron boundaries by PCR and directly sequenced the PCR products using the Applied Biosystems 3730 DNA analyzer (Thermo Fisher, USA).
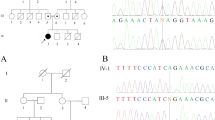
The insertion/deletion mutations were detected in 18 (36.7%) of 49 patients with RTT (Table 1). Among the 18 patients, 10 (55.6%) were considered novel by a comparison of our data with the known insertions/deletions deposited in the public databases, including RettBASE, gnomAD, Human Genome Mutation Database Professional 2019.2 and ClinVar. Representative data of pedigrees and sequences of the recombination breakpoints from three families are shown in Fig. 1. Patient 470 showed the insertion/deletion variant at c.1158_1258delinsCCGAGGGTGGCTCC. Patient 488 showed the deletion at c.1168_*539del. Patient 587 showed the insertion/deletion at c.1367_*791delinsCGC. Five (Patients 187, 470, 488, 559, and 587) had lost only exon 4. In addition, Patient 269 has a complex rearrangement with a 2609 bp deletion, including exon 3 and flanking introns, accompanied by two nucleotide substitution and a 25 bp deletion in exon4, c. [27-1707_c.378-206del; 1159_1160CC>AG; 1164_1188del], occurred in cis. The other five showed intragenic deletion involving exon 4 in the MECP2 locus. Hardwick et al. (2007) reported that 12 out of 21 patients (57%)4 experienced breakpoints within the “deletion-prone region (DPR)”, which is characterized by short direct repeat elements and is also known as the hotspot for the smaller deletions5,6. In this study, the breakpoints in seven novel insertion/deletion variants (7/10: 70%) were located within the DPR. In the breakpoints of Patients 269, 289 and 451, no repetitive sequences were found adjacent to the breakpoint. These findings suggest that the de novo deletion events involving MECP2 can be unique to families and that homology-mediated mechanisms are unlikely to be associated with these events.
In addition, we identified known pathogenic variants in 31 patients (Supplementary Table 1). No novel change was identified, suggesting that the molecular basis for recurrent de novo nucleotide substitutions in MECP2 is common among the different populations.
The list of MECP2 variants found in 49 Japanese patients with RTT should provide a useful resource to further examine the correlation between genotypes and disease phenotypes.
HGV Database
The relevant data from this Data Report are hosted at the Human Genome Variation Database at
https://doi.org/10.6084/m9.figshare.hgv.2648
https://doi.org/10.6084/m9.figshare.hgv.2651
https://doi.org/10.6084/m9.figshare.hgv.2654
https://doi.org/10.6084/m9.figshare.hgv.2657
https://doi.org/10.6084/m9.figshare.hgv.2660
https://doi.org/10.6084/m9.figshare.hgv.2663
https://doi.org/10.6084/m9.figshare.hgv.2666
https://doi.org/10.6084/m9.figshare.hgv.2669
https://doi.org/10.6084/m9.figshare.hgv.2672
https://doi.org/10.6084/m9.figshare.hgv.2675
https://doi.org/10.6084/m9.figshare.hgv.2678
https://doi.org/10.6084/m9.figshare.hgv.2681
https://doi.org/10.6084/m9.figshare.hgv.2684
https://doi.org/10.6084/m9.figshare.hgv.2687
https://doi.org/10.6084/m9.figshare.hgv.2690
https://doi.org/10.6084/m9.figshare.hgv.2693
https://doi.org/10.6084/m9.figshare.hgv.2696
https://doi.org/10.6084/m9.figshare.hgv.2699
https://doi.org/10.6084/m9.figshare.hgv.2702
https://doi.org/10.6084/m9.figshare.hgv.2705
https://doi.org/10.6084/m9.figshare.hgv.2708
https://doi.org/10.6084/m9.figshare.hgv.2711
https://doi.org/10.6084/m9.figshare.hgv.2714
https://doi.org/10.6084/m9.figshare.hgv.2717
https://doi.org/10.6084/m9.figshare.hgv.2720
https://doi.org/10.6084/m9.figshare.hgv.2723
https://doi.org/10.6084/m9.figshare.hgv.2726
https://doi.org/10.6084/m9.figshare.hgv.2729
https://doi.org/10.6084/m9.figshare.hgv.2732
https://doi.org/10.6084/m9.figshare.hgv.2735
https://doi.org/10.6084/m9.figshare.hgv.2738
https://doi.org/10.6084/m9.figshare.hgv.2741
https://doi.org/10.6084/m9.figshare.hgv.2744
https://doi.org/10.6084/m9.figshare.hgv.2747
https://doi.org/10.6084/m9.figshare.hgv.2750
https://doi.org/10.6084/m9.figshare.hgv.2753
https://doi.org/10.6084/m9.figshare.hgv.2756
https://doi.org/10.6084/m9.figshare.hgv.2759
https://doi.org/10.6084/m9.figshare.hgv.2762
https://doi.org/10.6084/m9.figshare.hgv.2765
https://doi.org/10.6084/m9.figshare.hgv.2768
https://doi.org/10.6084/m9.figshare.hgv.2771
https://doi.org/10.6084/m9.figshare.hgv.2774
https://doi.org/10.6084/m9.figshare.hgv.2777
References
Ip, J. P. K., Mellios, N. & Sur, M. Rett syndrome: insights into genetic, molecular and circuit mechanisms. Nat. Rev. Neurosci. 19, 368–382 (2018).
Amir, R. E. et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 23, 185–188 (1998).
Krishnaraj, R., Ho, G. & Christodoulou, J. RettBASE: Rett syndrome database update. Hum. Mutat. 38, 922–931 (2017).
Hardwick, SA., Reuter, K., Williamson, SL., Vasudevan, V., Donald, J. & Slater, K. Delineation of large deletions of the MECP2 gene in Rett syndrome patients, including a familial case with a male proband.Eur. J. Hum. Genet. 15, 1218–1229 (2007).
Laccone, F. et al. Large deletions of the MECP2 gene detected by gene dosage analysis in patients with Rett syndrome. Hum. Mutat. 3, 234–244 (2004).
Vidal, S. et al. Characterization of large deletions of the MECP2 gene in Rett syndrome patients by gene dosageanalysis. Mol. Genet. Genom. Med. 7, e793 (2019).
Acknowledgements
We are very grateful to the patients who participated in this study. We thank the attending physicians and Yoko Nakamura and Yoshie Sawano (Department of Mental Retardation and Birth Defect Research, National Institute of Neurology, NCNP) for their technical contribution. This study is partially supported by the Program for an Integrated Database of Clinical and Genomic Information (17kk0205012h0002 to Y.G.) and Construction of integrated database of clinical and genomics information and sustainable system for promoting genomic medicine in Japan (18kk0205012s0303 to Y.G.) from the Japan Agency for Medical Research and Development, AMED, and the Intramural Research Grant (27-6 to Y.G.; 30-9 to A.I. for Neurological and Psychiatric Disorders of NCNP, and Joint Usage and Joint Research Programs, the Institute of Advanced Medical Sciences, Tokushima University (A.I.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Takeshita, E., Iida, A., Abe-Hatano, C. et al. Ten novel insertion/deletion variants in MECP2 identified in Japanese patients with Rett syndrome. Hum Genome Var 6, 48 (2019). https://doi.org/10.1038/s41439-019-0078-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41439-019-0078-2