Abstract
Nitrate is a major nitrogen resource for plant growth and development and acts as both a crucial nutrient and a signaling molecule for plants; hence, understanding nitrate signaling is important for crop production. Abscisic acid (ABA) has been demonstrated to be involved in nitrate signaling, but the underlying mechanism is largely unknown in apple. In this study, we found that exogenous ABA inhibited the transport of nitrate from roots to shoots in apple, and the transcription of the nitrate transporter MdNRT1.5/MdNPF7.3 was noticeably reduced at the transcriptional level by ABA, which inhibited the transport of nitrate from roots to shoots. Then, it was found that the ABA-responsive transcription factor MdABI5 bound directly to the ABRE recognition site of the MdNRT1.5 promoter and suppressed its expression. Overexpression of MdABI5 inhibited ABA-mediated transport of nitrate from roots to shoots. Overall, these results demonstrate that MdABI5 regulates the transport of nitrate from roots to shoots partially by mediating the expression of MdNRT1.5, illuminating the molecular mechanism by which ABA regulates nitrate transport in apple.
Similar content being viewed by others
Introduction
Apple (Malus domestica) is an economically important fruit and is widely planted worldwide. Both genetic and environmental factors affect fruit quality and yield, and nutrient absorption and utilization are very important for apple growth. Nitrogen is the most important nutrient promoting crop productivity1. In agricultural production, effective utilization of nitrogen is the key to high plant yield, but as a great deal of nitrogen in the soil cannot be absorbed fully by plants, it is necessary to study the mechanisms and genes involved in nitrogen absorption, assimilation, and transport to improve nitrogen use efficiency.
NO3– and NH4 + are the major nitrogen sources in aerobic and anoxic soils, respectively2. For most crops, nitrate is the major nitrogen source3, and its uptake and transport by plants have been studied in depth. Nitrate is taken up into plants by nitrate transporters. To adapt to different environmental NO3− concentrations in the environment, plants have evolved two NO3− absorption systems, low-affinity transport systems, and high-affinity transport systems, with corresponding low affinity (NRT1) and high affinity (NRT2) NO3− transporters4. In Arabidopsis, the NRT1/PTR family contains 53 members, while the NRT2 family contains 7 members5,6. Some NRTs have also been functionally identified. For example, NRT1.1/CHL1 is both a nitrate sensor and dual-affinity nitrate transporter7,8. NRT1.1, NRT1.2, NRT2.1, and NRT2.2 are involved in nitrate uptake9,10,11, NRT1.4 regulates leaf nitrate homeostasis11, NRT1.6 participates in the transfer of nitrate from maternal tissue to developing embryos12, NRT1.7 regulates nitrate remobilization from source to sink tissue13, and NRT1.9 is involved in the transport of nitrate from roots to shoots14.
NRT1.5/NPF7.3 and NRT1.8 have been confirmed as two crucial nitrate transporters for long-distance transport15. NRT1.5 is expressed predominantly in root pericycle cells and plays an important role in loading nitrate into the xylem, while NRT1.8 is mainly expressed in xylem parenchyma cells and plays a major role in unloading nitrate from xylem sap10. Under normal conditions, NRT1.5 loads nitrate from roots into xylem vessels (from which it is transferred to aerial tissues), whereas NRT1.8 unloads nitrate into xylem parenchyma cells15. Microarray analysis demonstrated that NRT1.5 and NRT1.8 have opposite regulatory patterns under various abiotic and biotic stresses4. Nitrate reallocation to plant roots often occurs under stress; a previous study indicated that drought, cadmium, and salt treatments reduced the expression of NRT1.5, and the transcription of the abiotic stress-responsive genes Na+/H+ EXCHANGER1 (NHX1), SALTS OVERLY SENSITIVE1 (SOS1), RESPONSIVE TO DESICCATION29A (RD29A), and nitrate transporter NRT1.8 was upregulated in nrt1.5 mutant plants. These findings indicate that NRT1.5 may be involved in nitrate reallocation to regulate plant stress tolerance16.
In plants, nitrate is one of the most abundant anions, and its absorption and transport are believed to be accompanied by K+, NRT1.5 plays a crucial role in K+ translocation from roots to shoots and is also involved in the coordination of K+/ NO3− distribution in plants6,17. In addition to K+/NO3-, recent research has found that NRT1.5 transports IBA (indole-3-butyric acid) more efficiently than IAA, and IBA is the preferred substrate of NRT1.5 in vivo. NRT1.5-mediated IBA uptake into specific cells is involved in root gravitropism18.
Nitrate is both an important nutrient and signaling molecule for regulating the expression of many genes19. Nitrate inhibits anthocyanin accumulation primarily by suppressing the expression of production of anthocyanin pigments genes which are induced by light in Arabidopsis20. The bZIP transcription factor elongated hypocotyl 5 (HY5), which positively regulates plant photomorphogenesis, is involved in nitrogen signaling by promoting the expression of nitrate uptake genes and assimilation-related genes21. Nitrate signaling pathways for regulating phosphate utilization have been extensively studied. A previous study found that NRT1.1B could perceive nitrate signals and degrade the repressor protein SPX4, with subsequent release of core transcription factors for phosphate and nitrate signaling to ensure coordinated utilization of N and P in rice22. The NIN-like protein NLP7 is a central transcription factor that regulates the expression of NRT1.1 and NRT2.1 in Arabidopsis23. Ca2 + signaling is also involved in the response to nitrate24,25.
Various hormone signals affect plant nitrate contents. For example, the nitrate content of Arabidopsis plants changed when they were treated with ethylene and methyl jasmonate15. ABA is a crucial hormone that regulates many developmental processes and abiotic stress responses26. ABA has been demonstrated to be involved in nitrate signaling, and the expression levels of nitrate-related genes were altered under ABA treatment in Arabidopsis27. Nitrate provisioning following nitrate starvation increased root-tissue ABA accumulation in wheat, and this enhancement affected the expression of NRT2/NAR genes28. These findings demonstrate that there is some correlation between ABA and nitrate.
In this study, we examined the effect of exogenous ABA on the transport of nitrate in apple seedlings and investigated the pertinent regulatory mechanisms. We found that ABA treatment inhibited the transport of nitrate from roots to shoots, and the transcription factor MdABI5 bound directly to the promoter region of the nitrate transporter MdNRT1.5 and suppressed its expression. In brief, these results indicate that MdABI5 regulates root-to-shoot transport of nitrate by modulating the expression of MdNRT1.5, illuminating the molecular mechanism by which ABA regulates nitrate transport in apples.
Materials and methods
Plant materials and experimental treatments
“Orin” calli were subcultured on MS medium containing 1.5 mg L−1 6-benzylaminopurine (6-BA) and 0.5 mg L−1 2, 4-dichlorophenoxyacetic acid (2, 4-D) at 25 °C in the dark and subcultured at 18-day intervals. The “Royal Gala” apple cultivar was subcultured monthly at 25 °C, and the MS medium contained 1.5 mg L−1 6-BA, and 0.2 mg L−1 naphthylacetic acid. The wild-type Arabidopsis plants used were the Columbia (Col-0) type and were grown at 22 °C.
The basic nutrient solution contained 1.0 mM CaCl2, 1.0 mM KH2PO4, 1.0 mM MgSO4, 0.1 mM FeSO4·7H2O, 0.1 mM Na2EDTA·2H2O, 50 μM MnSO4⋅H2O, 50 μM H3BO3, 0.05 μM CuSO4⋅5H2O, 0.5 μM Na2MoO4⋅2H2O, 15 μM ZnSO4⋅7H2O, 2.5 μM KI, and 0.05 μM CoCl⋅6H2O, supplemented with 5 and 0.1 mM KNO3 as the sole N source, and the corresponding concentration of KCl was added to the nutrient solution to maintain the same K concentration (10 mM KCl). For ABA treatment, we exposed apple (Malus domestica “Royal Gala”) seedlings to high nitrogen (5 mM KNO3) nutrient solutions containing 1.5 mg L−1 6-BA, 0.2 mg L−1 naphthylacetic acid, and different concentrations of ABA (0 and 20 μM ABA) for 0, 1, 3, 6, and 12 h, and the gene expression of MdNRT1.5 was then measured. Apple calli were treated with high nitrogen (5 mM KNO3) medium containing 1.5 mg L−1 6-BA, 0.5 mg L−1 2, 4-D, and different concentrations of ABA (0 and 20 μM ABA) for one day, and then the samples were used for the GUS experiment.
The “Royal Gala” apple cultivar was planted in rooting medium (1/2 MS medium containing 0.2 mg L−1 IAA, pH = 5.8). After rooting, the seedlings were cultivated in basic nutrient solutions containing 5 mM KNO3 for 30 days, and then seedlings of similar size and status were transferred to vermiculite treated with different nitrate concentrations (5 mM and 0.1 mM KNO3) with different ABA concentrations (0, 5, and 20 μM ABA) for 50 days under long-day conditions (25 °C, 8/16-h dark/light). The treatment solutions were renewed every 7 days. After 50 days, the shoots and roots were separated, and we washed the roots with ddH2O, dried them with filter paper and measured the fresh weights and nitrate content. The shoots and roots were separated and dried at 80 °C for 3 d, and dry weights were measured.
Chinese crabapple (Malus hupenensis) seeds were stratified at 4 °C for 30 days. After germination, the seedlings were cultivated in 5 mM KNO3 nutrient solutions for 6 weeks, and then seedlings of similar size and status were transferred to vermiculite treated with different nitrate concentrations (5 mM and 0.1 mM KNO3) with different ABA concentrations (0, 5, and 20 μM ABA) for 50 days under a long-day condition (25 °C, 8/16-h dark/light). The treatment solutions were renewed every 7 days.
Arabidopsis seeds were sown and germinated on MS nutrient medium. After 1 week of growth, seedlings were transferred to different nitrate concentrations (5 mM and 0.1 mM KNO3) with different ABA concentrations (0 and 10 μM ABA). These Arabidopsis plants were grown under long-day conditions (22 °C, 8/16-h dark/light). The physiological data shown in Fig. S4 were evaluated after 14 days, and the physiological data shown in Fig. 6 were evaluated after 10 days.
Plasmid construction and plant transformation
The full-length cDNA sequences of MdNRT1.5 and MdABI5 were inserted into the pRI 101-GFP vector and the pCXSN-MYC vector to obtain MdNRT1.5-GFP and MYC-MdABI5 vectors, respectively, for overexpression analysis. These vectors were transformed into Arabidopsis and ‘Orin’ calli using Agrobacterium (GV4404)-mediated transformation.
To obtain MdABI5- and MdNRT1.5-overexpression apple roots, MdABI5 and MdNRT1.5 were inserted into the vector pK7GWIWG2 with a red fluorescent protein gene and the vector pRI 101-GFP with a green fluorescent protein, respectively. Then these constructs were subsequently transformed into apple seedlings roots with the same growth state as described by Zhao et al.29.
RNA extraction and qRT-PCR
RNA extraction and reverse transcription were carried out as described by Liu et al.30. Quantitative real-time polymerase chain reaction (qRT-PCR) was performed using Step One Plus. At least three biological replicates were analyzed for each sample. All primers used are listed in Supplementary Table S1.
Transcriptome analyses
Apple seedlings were grown on 0.1 mM KNO3 (the sole nitrogen source) for 5 days, half of the apple seedlings were treated with 0.1 mM KNO3 plus 20 μM ABA for 6 h, and the remaining apple seedlings were left to remain growing on 0.1 mM KNO3 medium for 6 h. Total RNA from plants treated with or without ABA was extracted and used for transcriptome analyses. The transcriptome was analyzed with the methods described by Zhang et al.31. Log2-fold change values >0.58 were considered significant. The significantly repressed (<0.58-fold) and induced (>1.5-fold) genes related to nitrate uptake and transport are shown in Fig. 2b, and the significantly expressed genes (|log2-fold change| > 0.58) are shown in Supplemental Appendix 1.
GUS-staining assay
To determine whether the transcriptional activity of the MdNRT1.5 promoter is reduced by ABA and verify that the ABRE core CACGTA sequence is vital for the ABA response, the normal and mutant promoters (ACAAAC in place of CACGTA) of MdNRT1.5 were cloned into the p1300-GN vector, which contains the GUS reporter gene. The resulting pMdNRT1.5::GUS and pMdNRT1.5::GUS (m) constructs were transiently transformed into apple calli. To verify the activation or suppression of MdNRT1.5 by MdABI5, 35S::MdABI5 was cotransformed into pMdNRT1.5::GUS calli. GUS staining and activity were used to determine the activity of the transgenic calli as described by Hu et al.32.
ChIP-PCR analyses
35S::MYC and 35S::MYC-MdABI5 transgenic calli were subjected to ChIP experiments. Apple calli were crosslinked with formaldehyde and sonicated to disrupt chromatin. An anti-MYC antibody (Beyotime) was used for Chromatin immunoprecipitation-PCR (ChIP-PCR) as described by Hu et al.33. qRT-PCR was performed to detect the enrichment of target DNA fragments. The primers used for ChIP-PCR are listed in Supplementary Table S1.
Electrophoretic mobility shift assays (EMSAs)
EMSA was conducted as described by Xie et al.34. MdABI5 was cloned into the vector pPET32a, The MdABI5-HIS recombinant proteins were expressed in and purified from Escherichia coli strain BL21 (DE3). The oligonucleotide probes of the MdNRT promoters were specificity labeled with 3’-biotin as described by Ma et al.35. The same unlabeled sequences were used as competitors. The primers used are listed in Supplemental Table S1.
Yeast one-hybrid (Y1H) assay
Y1H assays were performed as described by Wang et al.36. The coding sequence of MdABI5 was inserted into the pGADT7 expression vector, and the promoter fragment of MdNRT1.5 that contained the ABRE-binding motifs was inserted into the pHIS2 reporter vector. Then, constituent vectors were cotransformed into the yeast one-hybrid strain Y187. Individual transformants were screened on Trp-, Leu-, and His-deficient medium (SD-T/-L/-H), and then the positive yeast cells were identified by PCR. The positive yeast strain cells were diluted 10, 100, and 1000 times. Ten microliters of solution with different concentrations were dripped onto a selective medium (SD-T/-L/-H) supplemented with 50 mmol/L 3-aminotriazole (3-AT) for 2–3 days at 28 °C.
Nitrate content
The nitrate content was measured using the hydrazine reduction method as described by Liu et al.9. For nitrate content detection, apple seedlings were treated with nitrogen deficiency treatment for 5 days, and then the apple seedlings were treated with high nitrogen (5 mM KNO3) and low nitrogen (0.1 mM KNO3) nutrient solutions containing different concentrations of ABA (0, 5, and 20 μM). After a 50-d treatment, the roots and shoots of the apple seedlings were collected. Arabidopsis seeds were sown on MS medium. Four days after being sown, seedlings were transferred to different nitrate concentrations (5 and 0.1 mM KNO3) with different ABA concentrations (0 and 10 μM) after 10 d of treatment, and then the roots and shoots of Arabidopsis were collected. The samples were milled into powder, added to l ml ddH2O, and then boiled at 100 °C for 30 min. The samples were centrifuged for 10 min at 13,000 rpm, and then the supernatant was collected to a flow cell. The nitrate content was measured by using an AutoAnalyzer 3 continuous flow analytical system (SEAL Analytical).
Analysis of root-to-shoot nitrate allocation using 15NO3 −
WT and MdABI5-overexpressing apple plants were grown on basic nutrient solutions containing 5 mM KNO3 for 30 d and then treated with 20 μM ABA for 12 h. Plant material was washed in 0.1 mM CaSO4 for 1 min, labeled for 30 min in 5 mM K15NO3 or 0.1 mM K15NO3 medium (99% atom excess of 15N; pH 6.0), and washed again in 0.1 mM CaSO4 for 1 min. The shoots and roots were separated and dried at 80 °C for 3 d, and 15N-NO3− contents were analyzed using an isotope-ratio mass spectrometry system (Thermo Scientific, USA)15.
Statistical analysis
The data were obtained from three biological replicates. DPS software was used to analyze the significance of the differences as described by Zhang et al.37.
Results
Exogenous ABA inhibits the transport of nitrate from roots to shoots
To explore the possible effects of ABA on nitrogen uptake and transport, we exposed apple (M. domestica “Royal Gala”) seedlings to high nitrogen (5 mM KNO3) and low nitrogen (0.1 mM KNO3) nutrient solutions containing different concentrations of ABA (0, 5, and 20 μM). After a 50-d treatment, compared with the ABA-free control treatment, shoot growth was lower under ABA treatment (Fig. 1a). After ABA treatment, the fresh weights and dry weights of shoots and roots were clearly decreased (Fig. 1b–e), nitrate contents in both roots and shoots were reduced (Fig. 1f, g), and the shoot/total nitrate ratio was reduced substantially under ABA treatment (Fig. 1h), while the root/shoot nitrate ratio was increased substantially under ABA treatment (Fig. 1i). We performed the same treatments with Chinese crabapple (M. hupenensis) seedlings and obtained similar results (Fig. S1). These results indicated that exogenous ABA inhibited the transport of nitrate from roots to shoots in apple seedlings.
a Phenotype of apple (Malus domestica “Royal Gala”) seedlings after growth on high nitrogen (5 mM KNO3) and low nitrogen (0.1 mM KNO3) nutrient solutions containing different concentrations of ABA (0, 5, and 20 μM) for 50 d. b, c Fresh weights of shoots and roots after growth on 5 mM KNO3 and 0.1 mM KNO3 nutrient solutions containing different concentrations of ABA (0, 5, and 20 μM) for 50 d. d, e Dry weights of shoots and roots after growth on 5 mM KNO3 and 0.1 mM KNO3 nutrient solutions containing different concentrations of ABA (0, 5, and 20 μM) for 50 d. f, g Nitrate contents measured in shoots and roots after growth on 5 mM KNO3 and 0.1 mM KNO3 nutrient solutions containing different concentrations of ABA (0, 5, and 20 μM) for 50 d. h Shoot/total ratio of nitrate per apple seedlings. i Root/shoot ratio of nitrate per apple seedlings. Data were shown as means ± SE (n = 3). Student’s t test (*p < 0.05 and **p < 0.01) was used to analyze the statistical significance
ABA regulates the expression of genes involved in nitrogen uptake and transport
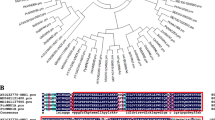
To understand the mechanism of nitrate regulation by ABA, we performed transcriptome analysis with and without ABA. Among the differentially expressed genes we identified [|Log2FC| > 0.58; p < 0.05], a total of 583 genes demonstrated altered expression (277 up, 306 down) (Fig. 2a; Supplemental Appendix 1); Compared with the levels in untreated controls (without ABA treatment), seven NRT/NPF genes demonstrated altered expression; among these genes, MdNPF4.5 (GenBank accession number: MD08G1040500), MdNRT2.4 (MD11G1141700), MdNRT1.5/MdNPF7.3 (MD06G1029400), MdNPF6.2 (MD15G1173800), MdNPF4.4 (MD05G1000900), and MdNPF4.3 (MD15G1443100) were notably reduced, while MdNPF5.4 (MD07G1039600) was significantly increased after ABA treatment (Fig. 2b). To further confirm the transcriptome results, we measured the expression of these genes by quantitative PCR (RT-qPCR). Compared with those in the untreated controls (without ABA treatment), the expression levels of MdNPF4.5, MdNRT2.4, MdNRT1.5, MdNPF6.2, MdNPF4.4, and MdNPF4.3 were rapidly reduced when apple seedlings were treated with ABA for 6 h (Fig. 2c). These results were consistent with the transcriptome data. NRT1.5 functions to load nitrate into the xylem to regulate nitrate long-distance transport from roots to shoots15. Phylogenetic analysis demonstrated that MdNRT1.5 clustered closely with AtNRT1.5 (Fig. S2a).
a Volcano map indicating up- and downregulation of differentially expressed genes [|Log2FC (0.1 mM KNO3 + 20 μΜ ABA/0.1 mM KNO3) | >0.58; p < 0.05] in response to ABA conditions. b Transcription levels of nitrate-related genes in seedling roots grown on low nitrate plates (0.1 mM KNO3) with or without 20 μM ABA treatment. c Quantitative detection of nitrate-related genes expression in the transcriptome
Considering that ABA inhibited nitrate transport from roots to shoots and that ABA repressed the transcription of MdNRT1.5, MdNRT1.5 was subsequently chosen for further investigation. We fused MdNRT1.5 to the expression vector pRI 101-GFP with a green fluorescent protein and transformed the construct into the roots of apple seedlings in the same growth state using the Agrobacterium rhizogenes-mediated transformation method29. We examined the fluorescence intensity and expression level of MdNRT1.5 in overexpression plants and obtained six MdNRT1.5-overexpressing apple roots with elevated MdNRT1.5 transcript levels (Fig. S3a, b). After 30 d of treatment with different concentrations of nitrate (5 mM KNO3 and 0.1 mM KNO3), no difference was found in the shoot/total nitrate ratio and root/shoot nitrate ratio between transgenic lines and WT under high nitrate (5 mM KNO3) treatment (Fig. S3e, f). However, the shoot/total nitrate ratio of the transgenic lines was higher and the root/shoot nitrate ratio was lower under the low nitrate (0.1 mM KNO3) treatment (Fig. S3e, f). We performed the same treatments with MdNRT1.5 transgenic Arabidopsis seedlings and obtained similar results (Fig. S4). These findings further suggested that the nitrate transporter MdNRT1.5 is able to transport nitrate from roots to shoots.
ABRE cis-elements in the promoter region of the MdNRT1.5 gene is required for ABA-mediated expression
Since the expression of MdNRT1.5 was inhibited by ABA, to further confirm the transcriptome results, the expression of MdNRT1.5 in apple seedlings treated with ABA was measured. The results indicated that the expression of MdNRT1.5 was remarkably reduced after 3 h of ABA treatment (Fig. 3a), further verifying that MdNRT1.5 expression was inhibited by ABA. We also analyzed the promoter of the MdNRT1.5 gene, which contains an ABA-responsive element (CACGTA). To explore whether the transcriptional activity of the MdNRT1.5 promoter was reduced by ABA, the MdNRT1.5 promoter was cloned into the p1300-GN vector, which contains the GUS reporter gene. A pMdNRT1.5::GUS vector was constructed and genetically transformed into apple calli. GUS assays revealed that ABA treatment dramatically reduced the GUS activity, demonstrating that the promoter of MdNRT1.5 was ABA-responsive (Fig. 3b, c).
To examine whether the ABA-responsive element (ABRE) core CACGTA sequence was vital for the ABA response, a mutated MdNRT1.5 promoter that contained ACAAAC in place of the wild-type core sequence was artificially constructed and placed in a GUS reporting vector (pMdNRT1.5::GUS (m)) as described previously. We also transformed this mutated construct into apple calli for GUS analysis. The GUS activity of pMdNRT1.5::GUS (m) transgenic calli was unchanged after ABA treatment (Fig. 3b, c), indicating that the ABRE cis-element in the MdNRT1.5 promoter region was critical to the ABA response.
MdABI5 binds directly to the promoter of MdNRT1.5 to repress its expression
The ABRE binding factor (ABF)/ABA-responsive element-binding protein (AREB) subfamily belongs to the bZIP transcription factors, which were reported to recognize and bind to ABRE cis-elements and regulate ABA-dependent gene expression, AREB proteins play an important role in the ABA response26, which motivated us to determine whether AREBs play a role in ABA-mediated regulation of nitrate. The expression of MdABI5 (LOC103430245), a homolog of AtABI5 (Fig. S2b), was noticeably induced by nitrate (Fig. S5). Therefore, MdABI5 was chosen for further study.
To verify whether MdABI5 is bound to the promoters of MdNRT1.5 and other MdNRTs, their promoters were analyzed, and several ABREs were found (Fig. S6a). We subsequently performed ChIP-PCR, which showed that the promoter regions of MdNRT1.5 and MdNRT2.4 were enriched in the 35::MYC-MdABI5 transgenic calli compared to the 35S::MYC control (Fig. 4a), indicating that MdABI5 might bind to the ABRE recognition sites of the MdNRT1.5 and MdNRT2.4 promoters. To determine whether MdABI5 is bound directly to the MdNRT1.5 and MdNRT2.4 promoters, we conducted an EMSA using MdABI5–HIS fusion protein, only an MdNRT1.5 DNA-MdABI5 protein complex was strongly detected (Fig. 4b), and the amount of complex decreased as the amount of the unlabeled ABRE competitor increased. When the ABRE cis-element CACGTA was subsequently mutated to ACAAAC, this competition was not observed as the amount of the unlabeled mutant competitor increased (Fig. 4c), indicating specific binding. MdABI5 is also bound directly to the AtNRT1.5 promoter (Fig. S6b).
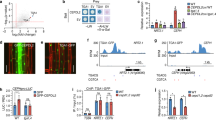
a ChIP-PCR assays of the enrichment of nitrate gene promoters in 35S::MYC and 35S::MYC-MdABI5 transgenic apple calli. The 18S served as the loading control. b EMSA of the interaction analysis between MdABI5 and labeled DNA probes in the promoters of MdNRT1.2, MdNRT1.5, MdNRT1.8, MdNRT1.9, MdNRT2.4, and MdNRT2.5. c The interaction between MdABI5 protein and the MdNRT1.5 promoter in the EMSA. Mutation probes of pMdNRT1.5 had a mutated ABRE where CACGTA was replaced with ACAAAC. d YIH assay of MdABI5 bound to the MdNRT1.5 promoter in vitro. 10−1, 10−2, and 10−3 indicate that the yeast concentration was diluted 10, 100, and 1000 times, respectively. 3-AT stands for 3-Amino-1, 2, 4-triazole
Yeast one-hybrid analysis was then conducted to further verify the interaction between MdABI5 and the MdNRT1.5 promoter. Cotransformed yeast cells harboring MdNRT1.5-P-pHIS2 and pGADT7 failed to grow on selective SD medium, but cells harboring MdNRT1.5-P-pHIS2 and MdABI5-pGADT7 grew well on selective medium (Fig. 4d), thereby demonstrating that MdABI5 bound directly to the MdNRT1.5 promoter.
To verify the suppression or activation of MdNRT1.5 by MdABI5, we tested the expression of MdNRT1.5 in MdABI5-overexpressing calli. The expression of MdNRT1.5 was decreased in MdABI5-overexpressing calli; in the presence of ABA, the expression of MdNRT1.5 was decreased further (Fig. 5a). The results indicated that MdABI5 repressed the expression of MdNRT1.5. To further verify this result, the 35S::MdABI5 constructs was transiently transformed into the aforementioned pMdNRT1.5::GUS transgenic calli. GUS activity analysis indicated that transgenic calli containing both pMdNRT1.5::GUS and 35S::MdABI5 exhibited much lower GUS activity than calli containing pMdNRT1.5::GUS alone. In the presence of ABA, GUS activity in these transgenic calli was lower, but calli containing both pMdNRT1.5::GUS and 35S::MdABI5 exhibited much lower GUS activity than those containing pMdNRT1.5::GUS alone (Fig. 5b, c). These results demonstrated that MdABI5 repressed the expression of MdNRT1.5 by binding directly to its promoter region.
a MdNRT1.5 expression levels in MdABI5-overexpressing calli with or without ABA treatments. b GUS staining experiment of pMdNRT1.5::GUS transgenic calli and transgenic calli containing both pMdNRT1.5::GUS and 35S::MdABI5 with or without ABA treatments. c GUS activity assays in transgenic apple calli
Ectopic expression of MdABI5 in Arabidopsis inhibits ABA-mediated transport of nitrate from roots to shoots
To identify whether MdABI5 was involved in the transport of nitrate from roots to shoots, 35S::MdABI5 was constructed and genetically transformed into Arabidopsis, and three independent MdABI5 transgenic lines with elevated transcription were obtained (Fig. S7a). MdABI5 transgenic Arabidopsis lines showed much lower levels of AtNRT1.5 transcription than untransformed controls (Fig. S7b). After ABA treatment, the transgenic lines’ fresh weight, root length, and nitrate content were lower than those of controls (Fig. 6 b-e, S8), their shoot/total nitrate ratio was reduced and root/shoot nitrate ratio was increased significantly under both low and high nitrogen conditions (Fig. 6f-i). These results indicated that MdABI5 inhibited ABA-mediated transport of nitrate from roots to shoots.
a Phenotype of MdABI5 transgenic lines and Col after growth on high nitrogen (5 mM KNO3) and low nitrogen (0.1 mM KNO3) nutrient solutions containing different concentrations of ABA (0 and 10 μM) for 10 d. b–e Nitrate content measured in various plant lines and organs (shoot and root) after growth on 5 mM KNO3 and 0.1 mM KNO3 nutrient solutions containing different concentrations of ABA (0 and 10 μM) for 10 d. f, g Shoot/total ratio of nitrate in various plant lines and organs (shoot and root). h, i Root/shoot ratio of nitrate in various plant lines and organs (shoot and root). Data were shown as means ± SE (n = 3). Student’s t test (*p < 0.05 and **p < 0.01) was used to analyze the statistical significance
MdABI5 enhances ABA-mediated repression of root-to-shoot nitrate transport
To test whether MdABI5 was involved in the response of ABA-repressed transport of nitrate in apple, we fused MdABI5 to the expression vector pK7GWIWG2 with an ERFP in its C-terminal region and transformed the construct into the roots of apple seedlings with the same growing state using the Agrobacterium rhizogenes-mediated transformation method29. We examined the fluorescence intensity and expression level of MdABI5 in overexpressed plants and obtained twelve MdABI5-overexpressing apple roots with elevated MdABI5 transcript levels (Figs. 7a and S7c). We then grew both WT and MdABI5-overexpressing plants in high nitrogen (5 mM KNO3) and low nitrogen (0.1 mM KNO3) nutrient solutions containing different concentrations of ABA (0 and 20 μM). After a 50-d treatment, the nitrate content, shoot/total nitrate ratio, and root/shoot nitrate ratio were measured (Figs. 7c–f and S9), and there was no difference in nitrate content, shoot/total nitrate ratio, or root/shoot nitrate ratio between WT and MdABI5-overexpressing plants under high nitrogen conditions (Figs. 7c, e and S9a). However, under low nitrogen conditions, the MdABI5-overexpressing plants had lower nitrate contents, lower shoot/total nitrate ratios, and higher root/shoot nitrate ratios than did the WT plants (Figs. 7d, f and S9c). Compared with the ABA-free treatment, ABA treatment reduced the nitrate content and shoot/total nitrate ratio and significantly increased the root/shoot nitrate ratio (Figs. 7c–f and S9). To further prove the function of MdABI5, WT, and MdABI5-overexpressing plants were treated with 0 and 20 μM ABA for 12 h and then labeled with different concentrations of K15NO3 (5 and 0.1 mM) for 30 min. Then the shoot/total nitrate (15NO3−) ratio and root/shoot nitrate (15NO3−) ratio were measured and similar results were obtained (Fig. 7g–j). These results indicated that MdABI5 enhanced ABA-mediated repression of nitrate transport from roots to shoots.
a RFP-red fluorescence intensity of apple roots. WT represents ‘Gala’ plants transformed with an empty vector containing an RFP tag; OE represents MdABI5-overexpressing apple roots. 1–12 represent different plant lines. b Phenotype of WT and MdABI5-overexpressing plants after growth on high nitrogen (5 mM KNO3) and low nitrogen (0.1 mM KNO3) nutrient solutions containing different concentrations of ABA (0 and 20 μM) for 50 d. c, d Shoot/total ratio of nitrate in various plant lines (WT and MdABI5-overexpressing plants) after growth on 5 mM KNO3 and 0.1 mM KNO3 nutrient solutions containing different concentrations of ABA (0 and 20 μM) for 50 d. e, f Root/shoot ratio of nitrate in various plant lines (WT and MdABI5-overexpressing plants). g, h WT and MdABI5-overexpressing plants were treated with 0 and 20 μM ABA for 12 h and then labeled with different concentrations of K15NO3 (5 mM and 0.1 mM) for 30 min. The shoot/total ratio of nitrate (15NO3−) was calculated. i, j The root/shoot ratio of nitrate (15NO3−) was calculated
Discussion
Phytohormone signaling molecules are involved in nitrate responses. Recent evidence has provided information concerning how the integration of nitrogen and phytohormone signals gives rise to changes in plant morphology and physiology. Among the various signals that regulate the expression of NRT genes, it has been proposed that auxins, cytokinins (CK), and ABA coordinate nitrogen acquisition12,38. For instance, AtNRT2.1 is induced by sugars and nitrate and repressed by CKs39. The evidence consistently indicates that CK is a root-to-shoot long-distance signal that represses transcription of AtNRT1.5, hence, it may be inferred that inhibition of AtNRT by CK reduces nitrate uptake activity27. Auxin may also mediate nitrogen uptake and transport because the expression of many NRT genes changes under IAA treatment40. In addition, the biosynthesis of CK and the biosynthesis and transport of auxin are affected by nitrate27. In addition, NRT1.1/CHL1 also promotes cell-to-cell auxin transport, providing another link between nitrate and auxin41.
There is some important evidence linking nitrogen and ABA in some plant species. Several findings provide genetic evidence that ABA is involved in lateral root development under high nitrate treatment in Arabidopsis27. Previous studies have shown that high-nitrate inhibition is significantly lower in ABA-deficient and ABA-insensitive mutants42. A previous study provided additional evidence for a link between nitrogen and ABA signaling in Medicago truncatula. A latd mutant (the LATD gene belongs to the NRT family and encodes a transporter) showed severe defects in root growth and root meristem maintenance that was rescued by providing exogenous ABA43. Another previous study demonstrated that knockdown of TabZIP60 (which is most closely related to the ABRE-binding factors AtABF2/AtAREB1, AtABF3, and AtABF4/AtAREB2) increased nitrogen uptake and grain yield, however, overexpression of TabZIP60 had the opposite effects44. Our study showed that treatment with exogenous ABA inhibited nitrate transport from roots to shoots in apple (Fig. 1) and reduced the transcription of multiple nitrate-related genes (Fig. 2b, c), providing evidence for a link between nitrate and ABA signaling.
In plants, K+/NO3− absorption, and transport are somehow coordinated and NRT1.5 is capable of transporting nitrate and potassium6. In addition, NRT1.5/NPF7.3 is an IBA transporter involved in root gravitropism18. Nitrate is both a crucial nutrient and a signaling molecule for plants. Many molecular components have been identified that are involved in nitrate signaling, including NLP6, NLP7, the TGA transcription factors TGA1 and TGA4, and squamosa promoter binding protein-like9 (SPL9)25,45,46. However, nitrate signaling is not clearly understood in apple and other fruit crops. MdABI5 expression was strongly induced after exposure to nitrate (Fig. S5). In addition to MdABI5, TaWabi5 in wheat (recognized as a putative ortholog of AtABI5) was also found to be nitrate inducible28, and ABI5 in Arabidopsis is involved in regulating C/N cross-talk and controlling lateral root development in response to nitrate44. In our study, phylogenetic analysis revealed that MdABI5 is closely related to MdAREB1 and MdAREB2 (Fig. S2b), again suggesting the importance of ABREs in nitrate signaling.
Nitrate reallocation to plant roots often occurs under stress conditions. Under stress, NRT1.5 expression is decreased in roots, inhibiting the transport of nitrate from roots to shoots and allowing nitrate accumulation in roots. This stress-initiated nitrate allocation to roots enhances plant stress tolerance47. Functional disruption of NRT1.5 enhances resistance to abiotic stresses; hence, these data indicate that stress-initiated downregulation of NRT1.5 leads to retention of nitrate in roots and improves plant stress tolerance16. Because exogenous or endogenous ABA generally stimulates jasmonic acid (JA) biosynthesis, it is believed that JA and ABA work synergistically in various biological processes48. JA downregulates the expression of NRT1.5 and inhibits nitrate transport from roots to shoots15. In our study, we found that (i) ABA-induced MdABI5 expression, (ii) MdABI5 inhibited the expression of MdNRT1.5 and regulated nitrate distribution in roots under ABA stress, and (iii) MdABI5 overexpression lines maintained a higher root nitrate proportion than WT when plants were treated with ABA. ABA-initiated nitrate allocation to roots would enable plants to respond and adapt to stress, enhancing plant stress tolerance. Thus, our data support the hypothesis that nitrate reallocation to roots is regulated by NRT1.5 genes and maybe a common stress response. It is possible that ABA and JA synergistically regulate nitrate reallocation, but we did not examine this possibility here.
After treatment with ABA, the expression levels of MdNRT2.4 and MdNRT1.5 were rapidly reduced (Fig. 2b, c), consistent with previous results15,27. Léran et al. found that the phosphatase ABI2 positively regulated NRT1.1/NPF6.3. In that study, ABA potentially reduced nitrate uptake by inactivating ABI2, which could be crucial for plant survival under stress conditions such as osmotic and drought stress, since nitrate absorption and utilization are energy-intensive processes49. These aforementioned studies demonstrate that ABA potentially represses nitrate uptake. In our study, we did not find direct binding of MdABI5 to the promoter of MdNRT2.4 in apple, whereas the expression of MdNPF4.5 was significantly decreased after ABA treatment (Fig. 2b, c). As OsNPF4.5 was found to promote the absorption of nitrate in rice50, we speculate that MdABI5 may regulate nitrate absorption by binding to the promoter of MdNPF4.5, but verifying this requires further study. Future work in this direction will not only improve our understanding of nitrate signaling but will also provide connections between the ABA and NO3− signaling pathways.
Overall, in this study, we found that MdABI5 was ABA-responsive and bound directly to the ABRE recognition site of the MdNRT1.5 promoter, repressing its expression and promoting ABA-mediated inhibition of nitrate transport from root to shoot. Our findings provide new insight into the molecular mechanism by which ABA regulates nitrate transport in apple.
References
He, X. et al. The nitrate-inducible NAC transcription factor TaNAC2-5A controls nitrate response and increases wheat yield. Plant Physiol. 169, 1991–2005 (2015).
Loque, D., Tillard, P., Gojon, A. & Lepetit, M. Gene expression of the NO3- transporter NRT1.1 and the nitrate reductase NIA1 is repressed in Arabidopsis roots by NO2-, the product of NO3- reduction. Plant Physiol. 132, 958–967 (2003).
Zhao, L., Liu, F., Crawford, N. M. & Wang, Y. Molecular regulation of nitrate responses in plants. Int. J. Mol. Sci. https://doi.org/10.3390/ijms19072039 (2018).
Li, J. Y. et al. The Arabidopsis nitrate transporter NRT1.8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance. Plant Cell 22, 1633–1646 (2010).
Leran, S. et al. A unified nomenclature of nitrate transporter 1/peptide transporter family members in plants. Trends Plant Sci. 19, 5–9 (2014).
Li, H. et al. NRT1.5/NPF7.3 functions as a proton-coupled H+/K+ antiporter for K+ loading into the xylem in Arabidopsis. Plant Cell 29, 2016–2026 (2017).
Hu, B. et al. Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat. Genet. https://doi.org/10.1038/ng.3337 (2015).
Xu, N. et al. The Arabidopsis NRG2 protein mediates nitrate signaling and interacts with and regulates key nitrate regulators. Plant Cell 28, 485–504 (2016).
Liu, F. et al. The long noncoding RNA T5120 regulates nitrate response and assimilation in Arabidopsis. N. Phytol. 224, 117–131 (2019).
Lin, S. H. et al. Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell 20, 2514–2528 (2008).
Chiu, C. C. et al. Mutation of a nitrate transporter, AtNRT1:4, results in a reduced petiole nitrate content and altered leaf development. Plant Cell Physiol. 45, 1139–1148 (2004).
Almagro, A., Lin, S. H. & Tsay, Y. F. Characterization of the Arabidopsis nitrate transporter NRT1.6 reveals a role of nitrate in early embryo development. Plant Cell 20, 3289–3299 (2008).
Liu, W. W., Sun, Q., Wang, K., Du, Q. G. & Li, W. X. Nitrogen limitation adaptation (NLA) is involved in source-to-sink remobilization of nitrate by mediating the degradation of NRT1.7 in Arabidopsis. N. Phytol. 214, 734–744 (2017).
Wang, Y. Y. & Tsay, Y. F. Arabidopsis nitrate transporter NRT1.9 is important in phloem nitrate transport. Plant Cell 23, 1945–1957 (2011).
Zhang, G. B., Yi, H. Y. & Gong, J. M. The Arabidopsis ethylene/jasmonic acid-NRT signaling module coordinates nitrate reallocation and the trade-off between growth and environmental adaptation. Plant Cell 26, 3984–3998 (2014).
Chen, C. Z., Lv, X. F., Li, J. Y., Yi, H. Y. & Gong, J. M. Arabidopsis NRT1.5 is another essential component in the regulation of nitrate reallocation and stress tolerance. Plant Physiol. 159, 1582–1590 (2012).
Du, X. Q. et al. The transcription factor MYB59 regulates K(+)/NO3 (-) translocation in the arabidopsis response to low K(+) stress. Plant Cell 31, 699–714 (2019).
Watanabe, S. et al. The Arabidopsis NRT1/PTR FAMILY protein NPF7.3/NRT1.5 is an indole-3-butyric acid transporter involved in root gravitropism. Proc. Natl Acad. Sci. USA 117, 31500–31509 (2020).
Wang, X. F. et al. The nitrate-responsive protein MdBT2 regulates anthocyanin biosynthesis by interacting with the MdMYB1 transcription factor. Plant Physiol. 178, 890–906 (2018).
Rubin, G., Tohge, T., Matsuda, F., Saito, K. & Scheible, W. R. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 21, 3567–3584 (2009).
An, J. P. et al. The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hortic. Res. 4, 17023 (2017).
Hu, B. et al. Nitrate-NRT1.1B-SPX4 cascade integrates nitrogen and phosphorus signalling networks in plants. Nat. Plants 5, 401–413 (2019).
Yu, L. H. et al. Overexpression of Arabidopsis NLP7 improves plant growth under both nitrogen-limiting and -sufficient conditions by enhancing nitrogen and carbon assimilation. Sci. Rep. 6, 27795 (2016).
Hu, H. C., Wang, Y. Y. & Tsay, Y. F. AtCIPK8, a CBL-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. Plant J. 57, 264–278 (2009).
Zhao, L. et al. The Arabidopsis NLP7 gene regulates nitrate signaling via NRT1.1-dependent pathway in the presence of ammonium. Sci. Rep. 8, 1487 (2018).
Ma, Q. J. et al. An apple CIPK protein kinase targets a novel residue of AREB transcription factor for ABA-dependent phosphorylation. Plant Cell Environ. 40, 2207–2219 (2017).
Kiba, T., Kudo, T., Kojima, M. & Sakakibara, H. Hormonal control of nitrogen acquisition: roles of auxin, abscisic acid, and cytokinin. J. Exp. Bot. 62, 1399–1409 (2011).
Wang, M. et al. TaANR1-TaBG1 and TaWabi5-TaNRT2s/NARs link ABA metabolism and nitrate acquisition in wheat roots. Plant Physiol. 182, 1440–1453 (2020).
Zhao, X. Y. et al. The MdWRKY31 transcription factor binds to the MdRAV1 promoter to mediate ABA sensitivity. Hortic. Res. https://doi.org/10.1038/s41438-019-0147-1 (2019).
Liu, Y. J. et al. MdBZR1 regulates ABA response by modulating the expression of MdABI5 in apple. Plant Cell Rep. 40, 1127–1139 (2021).
Zhang, J. C. et al. Phosphate regulates malate/citrate-mediated iron uptake and transport in apple. Plant Sci. https://doi.org/10.1016/j.plantsci.2020.110526 (2020).
Hu, D. G. et al. The R2R3-MYB transcription factor MdMYB73 is involved in malate accumulation and vacuolar acidification in apple. Plant J. 91, 443–454 (2017).
Hu, D. G., Sun, C. H., Zhang, Q. Y., Gu, K. D. & Hao, Y. J. The basic helix-loop-helix transcription factor MdbHLH3 modulates leaf senescence in apple via the regulation of dehydratase-enolase-phosphatase complex 1. Hortic Res. https://doi.org/10.1038/s41438-020-0273-9 (2020).
Xie, X. B. et al. The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ. 35, 1884–1897 (2012).
Ma, Q. J. et al. Transcription factor AREB2 is involved in soluble sugar accumulation by activating sugar transporter and amylase genes. Plant Physiol. 174, 2348–2362 (2017).
Wang, F. P., Wang, X. F., Zhang, J. C., Ma, F. W. & Hao, Y. J. MdMYB58 modulates Fe homeostasis by directly binding to the MdMATE43 promoter in plants. Plant Cell Physiol. 59, 2476–2489 (2018).
Zhang, Q. Y. et al. BTB-BACK-TAZ domain protein MdBT2-mediated MdMYB73 ubiquitination negatively regulates malate accumulation and vacuolar acidification in apple. Hortic Res. https://doi.org/10.1038/s41438-020-00384-z (2020).
Argueso, C. T., Ferreira, F. J. & Kieber, J. J. Environmental perception avenues: the interaction of cytokinin and environmental response pathways. Plant Cell Environ. 32, 1147–1160 (2009).
Brenner, W. G., Romanov, G. A., Kollmer, I., Burkle, L. & Schmulling, T. Immediate-early and delayed cytokinin response genes of Arabidopsis thaliana identified by genome-wide expression profiling reveal novel cytokinin-sensitive processes and suggest cytokinin action through transcriptional cascades. Plant J. 44, 314–333 (2005).
Fukaki, H. & Tasaka, M. Hormone interactions during lateral root formation. Plant Mol. Biol. 69, 437–449 (2009).
Krouk, G. et al. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell 18, 927–937 (2010).
Zhang, H., Rong, H. & Pilbeam, D. Signalling mechanisms underlying the morphological responses of the root system to nitrogen in Arabidopsis thaliana. J. Exp. Bot. 58, 2329–2338 (2007).
Yendrek, C. R. et al. A putative transporter is essential for integrating nutrient and hormone signaling with lateral root growth and nodule development in Medicago truncatula. Plant J. 62, 100–112 (2010).
Yang, J. B. et al. Reducing expression of a nitrate-responsive bZIP transcription factor increases grain yield and N use in wheat. Plant Biotechnol. J. 17, 1823–1833 (2019).
Krouk, G., Mirowski, P., LeCun, Y., Shasha, D. E. & Coruzzi, G. M. Predictive network modeling of the high-resolution dynamic plant transcriptome in response to nitrate. Genome Biol. 11, R123 (2010).
Alvarez, J. M. et al. Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. Plant J. 80, 1–13 (2014).
Yuan, J. Z. et al. ZxNPF7.3/NRT1.5 from the xerophyte Zygophyllum xanthoxylum modulates salt and drought tolerance by regulating NO3-, Na+ and K+ transport. Environ. Exp. Bot. https://doi.org/10.1016/j.envexpbot.2020.104123 (2020).
Wang, Y. F. et al. Abscisic acid promotes jasmonic acid biosynthesis via a ‘SAPK10-bZIP72-AOC’ pathway to synergistically inhibit seed germination in rice (Oryza sativa). New Phytol. https://doi.org/10.1111/nph.16774 (2020).
Leran, S. et al. Nitrate sensing and uptake in Arabidopsis are enhanced by ABI2, a phosphatase inactivated by the stress hormone abscisic acid. Sci. Signal. 8, ra43 (2015).
Wang, S. et al. Functional analysis of the OsNPF4.5 nitrate transporter reveals a conserved mycorrhizal pathway of nitrogen acquisition in plants. Proc. Natl Acad. Sci. USA 117, 16649–16659 (2020).
Acknowledgements
We sincerely thank our team leader Dr. Yu-Jin Hao. He will be remembered for his great achievements and for supporting and aiding in our work. This work was supported by grants from the National Key R&D Program of China (2018YFD1000100), National Natural Science Foundation of China (31772288 and 31972378), China Agriculture Research System of MOF and MARA (CARS-27), and the Agricultural Variety Improvement Project of Shandong Province (2019LZGC007).
Author information
Authors and Affiliations
Contributions
C.-X.Y., X.-F.W., Y.-J.L., and Y.-J.H. planned and designed the research. Y.-J.L., N.G., Q.-J.M., J.-C.Z., X.W., and J.L. performed the experiments and analyzed the data. Y.-J.L., X.-F.W., and C.-X.Y. wrote the paper.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, YJ., Gao, N., Ma, QJ. et al. The MdABI5 transcription factor interacts with the MdNRT1.5/MdNPF7.3 promoter to fine-tune nitrate transport from roots to shoots in apple. Hortic Res 8, 236 (2021). https://doi.org/10.1038/s41438-021-00667-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41438-021-00667-z