Abstract
Downy mildew (DM), caused by obligate parasitic oomycetes, is a destructive disease for a wide range of crops worldwide. Recent outbreaks of impatiens downy mildew (IDM) in many countries have caused huge economic losses. A system to reveal plant–pathogen interactions in the early stage of infection and quickly assess resistance/susceptibility of plants to DM is desired. In this study, we established an early and rapid system to achieve these goals using impatiens as a model. Thirty-two cultivars of Impatiens walleriana and I. hawkeri were evaluated for their responses to IDM at cotyledon, first/second pair of true leaf, and mature plant stages. All I. walleriana cultivars were highly susceptible to IDM. While all I. hawkeri cultivars were resistant to IDM starting at the first true leaf stage, many (14/16) were susceptible to IDM at the cotyledon stage. Two cultivars showed resistance even at the cotyledon stage. Histological characterization showed that the resistance mechanism of the I. hawkeri cultivars resembles that in grapevine and type II resistance in sunflower. By integrating full-length transcriptome sequencing (Iso-Seq) and RNA-Seq, we constructed the first reference transcriptome for Impatiens comprised of 48,758 sequences with an N50 length of 2060 bp. Comparative transcriptome and qRT-PCR analyses revealed strong candidate genes for IDM resistance, including three resistance genes orthologous to the sunflower gene RGC203, a potential candidate associated with DM resistance. Our approach of integrating early disease-resistance phenotyping, histological characterization, and transcriptome analysis lay a solid foundation to improve DM resistance in impatiens and may provide a model for other crops.
Similar content being viewed by others
Introduction
Downy mildew (DM) is a destructive disease caused by obligate parasitic oomycetes from the Peronosporaceae family1. It has been a serious challenge for a wide range of cultivated crops including row crops, vegetables, fruits, and ornamental plants. DM is globally distributed and has high adaptability to new and changing environmental conditions2. Most DM pathogens can infect their host plant at the seedling stage, causing systemic shoot infection, whereas infection at a more mature stage may develop into localized infection patches3. DM can affect the leaves, flowers, fruits, and shoots of hosts and cause great economic losses. It may lead to yield losses of up to 40–80% for different crops4,5. Many fungicides have been developed to manage DM pathogens; however, due to genetic recombination, frequent mutations, and asexual reproduction, new DM pathogen races with higher virulence levels emerge constantly, resulting in fungicide resistance in DM pathogens and thus severely hindering the effectiveness of fungicides whose development could take many years and cost hundreds of millions of dollars6,7.
DM pathogens are composed of at least 300 species belonging to different genera, such as Peronospora, Pseudoperonospora, and Plasmopara, among which Peronospora is the largest genus containing more than 260 species8. The common DM species infecting horticultural crops include Peronospora destructor (onion), Peronospora belbahrii (basil), Plasmopara viticola (grape), Pseudoperonospora cubensis (cucurbits), Plasmopara halstedii (sunflower), Peronospora effusa (spinach), and Bremia lactucae (lettuce). To combat this disease, host resistance to DM has been identified in several crops and a few resistance genes have been cloned. For example, the sunflower genome contains more than 30 DM resistance genes distributed in the domesticated and wild species9. In lettuce, over 50 DM resistance genes have been identified and genetically characterized, among which at least 28 genes can provide high levels of resistance against DM10. In grapevine, 27 quantitative trait loci (QTLs) for DM resistance have been identified from various Vitis species, of which the locus Rpv3 is a major determinant for DM resistance11. DM pathogens secrete apoplastic and cytoplasmic effector molecules upon infection that can be recognized by the proteins encoded by plant disease-resistance genes (R-genes), which are primarily comprised of nucleotide-binding site leucine-rich repeat (NBS-LRR) genes. Many NBS-LRR clusters have been identified in sunflower and lettuce genomic regions involved in DM resistance12,13. In Arabidopsis, some Toll/interleukin-1 receptor NBS-LRR (TIR-NBS-LRR) genes such as RPP1 confer organ-specific resistance to downy mildew14. In spinach, NBS genes present at the RPF1 locus contribute to resistance to P. effusa15.
Impatiens are one of the top-selling annual bedding flowers in the United States. The genus Impatiens (family Balsaminaceae) contains >1000 species that are widely distributed in different geographic and climatic regions, including tropical Africa, Southeast Asia, parts of Europe, and North America16. Among these species, Impatiens walleriana and Impatiens hawkeri are the most commonly cultivated in the world. The popularity of impatiens in the floriculture industry is attributed to the flower color diversity, profuse flowering nature, and ease of growing17,18. In 2018 alone, impatiens contributed a wholesale value of more than $109 million19. Impatiens downy mildew (IDM) caused by Plasmopara obducens is currently a huge threat to the impatiens industry20. Severe outbreaks of IDM were reported in Europe21, Australia22, and North America23,24, causing significant economic losses. The outbreak of IDM in the USA has caused a significant decrease of the wholesale values of impatiens from ~$150 million in 2005 down to ~$65 million in 201525. IDM caused by P. obducens has become a major disease of I. walleriana. The infected plants exhibit downward leaf curling, chlorotic and downy leaves, and leaves and flowers drop, all of which may result in complete losses of the aesthetic value of impatiens cultivars24. Several studies reported the morphology, transmission and hosts of P. obducens. This pathogen develops hyaline and monopodial sporangiophores with apical branches that can produce ovoid and hyaline sporangia26. The pathogen is readily transmitted by wind-blown or water-splashed sporangia from which the zoospores can be released and infect impatiens under suitable temperature and relative humidity27. Usually, 5–14 days after pathogen infection, visible white downy symptoms could be observed on the lower leaf surfaces27,28. The oospores could not be observed in fresh leaves and may survive overwinter in plant debris26,27,29. Plasmopara obducens can infect a number of cultivated and wild Impatiens spp., including I. walleriana, I. balsamina, I. pallida, I. carpensis, and I. glandulifera28,30,31,32,33. However, I. hawkeri appears to be highly resistant to this disease22.
Management of IDM can be achieved by using preventive fungicides. Several fungicides have been used to manage this disease in impatiens production facilities. Frequent applications of these fungicides have significantly increased the production costs and caused serious concerns over pesticide pollution of the environment. Moreover, few fungicides are available for use to manage this disease in the landscape (public or residential) and indoor exhibitions where impatiens are grown34,35. Developing and using disease-resistant cultivars have proven to be an effective, economic, and sustainable approach to managing devastating diseases in crops if genetic disease resistance can be found or developed. For example, disease-resistant cultivars have played an essential and critical role in controlling grapevine DM caused by P. viticola, which is in the same genus with the IDM pathogen P. obducens36. To develop disease-resistant cultivars, disease screening is essential and most critical. First, germplasm accessions, as many as possible, need to be screened to discover useful sources of disease resistance. Then, large breeding populations, generation after generation, need to be screened to identify the resistant progeny. Thus, effective and efficient disease screening or resistance phenotyping techniques frequently determine the success of plant disease-resistance breeding in many crops. IDM resistance has become the most important breeding objective in impatiens in the world; the development of effective and efficient IDM screening techniques would be of tremendous value to this important crop.
RNA-sequencing (RNA-Seq) technology has been used to identify genes potentially involved in DM resistance in horticultural plants, including lettuce, grapevine, spinach, and impatiens37,38,39,40,41,42. Previously, two de novo RNA-Seq comparative analyses of impatiens have identified some differentially expressed genes, including a couple of NBS-LRR genes for IDM resistance and candidates for IDM susceptibility41,42. However, in the absence of a reference genome, an accurate transcriptome and full lengths of the candidate genes are hardly achievable using short reads of RNAs based on the Illumina sequencing platform. Isoform sequencing (Iso-Seq), an advanced technique based on the single-molecule real-time (SMRT) sequencing platform and long reads of RNAs, has facilitated retrieval of full-length transcripts, assembly of high-quality reference transcriptomes, and discovery of splicing events and novel transcripts43. With Iso-Seq, each mRNA-derived cDNA molecule in a transcriptome is sequenced multiple rounds, resulting in high-quality full-length cDNA or corresponding mRNA sequences. Currently, a reference-level high-quality transcriptome for impatiens is not available. Study of the gene expression profiles at different developmental stages for different resistant and susceptible plants can function as a model to study DM-plant interaction at the transcriptome level and to uncover the plant–pathogen dynamics during resistance development.
In this study, the disease responses of 32 impatiens cultivars, including 16 I. walleriana and 16 I. hawkeri cultivars, were investigated at the cotyledon, first/second pair of true leaf, and mature plant stages, aiming to establish a system for early and rapid screening and phenotyping of impatiens for IDM resistance. DM pathogen growth and development in cotyledons and leaves were examined histologically, revealing the IDM-resistance mechanisms in I. hawkeri. Moreover, full-length transcriptome sequencing combined with RNA-Seq was applied to investigate transcriptome dynamics for three representative cultivars showing different resistance and susceptibility at cotyledon and true leaf stages. The transcriptome comparisons between IDM-resistant and susceptible cultivars and tissues revealed a core set of genes, including three R-genes potentially involved in IDM resistance in impatiens. Results from this study have provided very useful genomic resources and laid a solid foundation for future studies to implement genomics-assisted breeding of impatiens for IDM resistance and to identify and clone the IDM-resistance genes in impatiens in the future.
Results
Responses of 32 cultivars to natural downy mildew pathogen infection
In total, 16 cultivars of I. walleriana and 16 cultivars of I. hawkeri (Table 1) were evaluated in the field for their response to IDM at the mature stage. On December 28, 2014 (206 days after planting (DAP)) (average temperature 20.51 °C, relative humidity 88%, rainfall 0 cm)44, “Balance Orange” (BO) and “Super Elfin Pink” (SEP) of I. walleriana first showed IDM white sporulation on the abaxial side of foliage. Within 3 days, all plants of I. walleriana, sooner or later, showed similar IDM symptoms (Table 1). Infected impatiens plants showed chlorotic and downward-curling leaves, followed by leaf and flower dropping, complete defoliation, and plant collapse within a 7-week period. All plants died before February 16, 2015 (256 DAP), indicating all these I. walleriana cultivars are highly susceptible to IDM. By contrast, all plants of I. hawkeri cultivars did not show any IDM disease symptoms through the field experiment, suggesting that they possess strong resistance to IDM at the mature plant stage (Table 1).
All infected I. walleriana plants in the field showed chlorotic and downward-curling leaves with white downy mildew sporulation (growth) on the lower surface at the early infection stage and then followed by leaf and flower drops and plant collapsing in a seven-week period. No disease symptom was observed on I. hawkeri plants during field experiments. Details of disease incidence for inoculation experiments were described in Supplementary Table S1. “S” indicates susceptibility to impatiens downy mildew; “R” indicates resistance to impatiens downy mildew.
Phenotyping for downy mildew resistance at the earliest plant growth stages
To develop an effective early and rapid phenotyping system, we inoculated young plants of these 32 cultivars at their earliest growth stages (cotyledon and first/second pair of true leaf stages) as well as mature leaves using two inoculation methods (P. obducens spores applied to the abaxial or adaxial side of the cotyledons, first/second pair of true leaves, and mature leaves). Results showed that all I. walleriana cultivars were highly susceptible to IDM at all these stages (Table 1 and Supplementary Table S1). Typical white downy mildew sporulation was evident and profuse on the abaxial side of cotyledons and true leaves for all I. walleriana cultivars (Fig. 1A, D). All 16 cultivars of I. hawkeri showed resistance to IDM at the first/second pair of true leaf stage (Fig. 1E, F), consistent with typical plant responses of these cultivars to IDM at the mature stage. These results indicate that young impatiens plants at their first true leaf stage have developed resistance to IDM and are ready for IDM disease screening or phenotyping for IDM resistance.
Photos were taken 10 days post inoculation. A Cotyledon of I. walleriana cv. “Super Elfin Red”; B cotyledon of I. hawerki cv. “Divine Orange Bronze Leaf”; C cotyledon of I. hawerki cv. “Florific Lavender”; D second true leaf of I. walleriana cv “Super Elfin Red”; E second true leaf of I. hawerki cv. “Divine Orange Bronze Leaf”; F second true leaf of I. hawerki cv. “Florific Lavender”; and G second true leaf of I. hawerki cv. “Divine White Blush”. The red circles highlight the white downy mildew sporulation
Interestingly, I. hawkeri plants at the cotyledon stage exhibited different responses to inoculated P. obducens spores (P < 0.05). When inoculated on the abaxial side of cotyledons, “Divine Orange Bronze Leaf” (DOB) (Fig. 1B), “Divine Burgundy” (DB), “Divine Orange” (DO), and “Florific Violet” (FV) were susceptible to IDM with a disease incidence index at 0.67, 0.61, 0.56, and 0.50 at 10 days post inoculation (dpi), respectively (Supplementary Table S1). “Florific Sweet Orange” (FSO), “Divine White Blush” (DWB), “Florific White” (FW), and “Divine Violet” (DV) were also susceptible to DM, but with a lower disease incidence index (≤0.14). Most importantly, “Florific Lavender” (FLR) (Fig. 1C) and “Divine Lavender” (DL) showed strong resistance to IDM even at this early stage. There were no significant differences between 10 and 20 dpi across all cultivars, except that “Divine Pink” (DP) and DO showed significant differences when inoculated on the abaxial and adaxial side, respectively (Supplementary Table S1). When the adaxial side was inoculated, the disease incidence index was lower than that of the abaxial side (P < 0.05) for most cultivars except DV, FW (higher at the adaxial side). Again, DL and FLR showed strong resistance to IDM after their cotyledons were inoculated on either side, with zero disease incidence (Supplementary Table S1). On the other hand, DOB and DB consistently showed susceptibility to IDM when either side of the cotyledon was inoculated. These results indicated that 14 I. hawkeri cultivars were susceptible to IDM at the cotyledon stage and turned resistant starting at the true leaf stage, while two I. hawkeri cultivars (DL and FLR) expressed strong resistance to IDM starting at the cotyledon stage.
An interesting feature was observed on the adaxial and abaxial surfaces of inoculated cotyledons of I. hawkeri, but not on cotyledons of I. walleriana cultivars. During incubation after inoculation with P. obducens spores, irregular black “spots” and “specks” began to develop on cotyledon surfaces. Their occurrence varied among I. hawkeri cultivars but seemed to be from necrotic cells. For simplicity and convenience, we tentatively called them as “black spots”. To quantify the severity of black spots on cotyledons, we developed a black spot severity scale and calculated a black spot severity index (Fig. 2 and Supplementary Table S2). Black spot severity index for all I. hawkeri cultivars, except for “Divine Blue Pearl” (DBP), seemed to remain unchanged from 10 to 20 dpi. The cultivars DOB and DWB exhibited a higher black spot severity index than other cultivars at 10 dpi, with an average index of 2.18 and 2.08, respectively (Supplementary Table S2). Five cultivars, including FSO, FW, FLR, “Divine Cherry Red” (DCR), and DP showed a lower black spot severity index (<0.60). A similar trend was observed on the adaxial side of inoculated cotyledons, except that black spot severity was generally lower. The black spot severity index at 10 dpi appeared to be less than that at 20 dpi, but significant differences were not detected. At the first and second pair of true leaf stages, all I. hawkeri cultivars showed resistance to IDM and had no black spots, except DWB displaying small black spots on the leaf surface (Fig. 1G). When the IDM disease incidence indices (Supplementary Table S1) and the black spot severity indices of the 16 I. hawkeri cultivars (Supplementary Table S2) were examined, the Pearson Correlation Coefficient was 0.66 (abaxial, 10 dpi), 0.53 (abaxial, 20 dpi), 0.50 (adaxial, 10 dpi), and 0.51 (adaxial, 20 dpi), respectively, with an average of 0.55. Therefore, in general, the IDM disease incidence index had a moderate level of positive relationship with black spot severity. For example, the cotyledons of DOB exhibited high IDM incidence and severe black spotting after inoculation with P. obducens. However, there were some exceptions, for example, the cotyledons of DWB showed low IDM incidence yet severe black spotting. These results show that impatiens plants at their cotyledon stage may not express their typical mature plant resistance to P. obducens. However, if they do express resistance at this stage, they retain the resistance to IDM through their growth and developmental stages.
Based on the coverage of black spots on the inoculated cotyledon, black spot severity was scored in a scale of 0 to 4. A scale 0, no black spots; B scale 1, black spots covering less than ¼ of the cotyledon; C scale 2, black spots covering ¼ to ½ of the cotyledon; D scale 3, black spots covering ½ to ¾ of the cotyledon; and E scale 4, black spots covering more than ¾ of the cotyledon
Histological characterization of the disease-resistance response
Three cultivars, I. walleriana SER, I. hawkeri DOB and FLR, were selected for detailed histological characterization. In the above-described phenotyping experiments, they showed contrasting IDM-resistance responses: SER—susceptible at cotyledon, first/second pair of true leaf, and mature plant stages; DOB—susceptible at the cotyledon stage and resistant at the first/second pair of true leaf and mature plant stages; and FLR—resistant at cotyledon, first/second true leaf, and mature plant stages. Their cotyledons and true leaves were excised, inoculated with P. obducens sporangia, and cultured on 1% water agar. White downy mildew sporulation was evidently observed on the abaxial surface of cotyledons of SER and DOB at 4 dpi and became more and more massive at 6, 8, and 10 dpi. By contrast, only tiny mildew sporulation was observed on the cotyledons of FLR until 8 or 10 dpi, and the area of DM was very limited and not enlarging. When true leaves were inoculated, white mildew sporulation was only observed on the abaxial leaf surface of SER at 6 dpi but not on leaf surfaces of DOB and FLR. These results confirmed that the cotyledons of SER and DOB and true leaves of SER were susceptible to IDM, while the cotyledons of FLR and true leaves of DOB and FLR were resistant to IDM.
The sporangia density on the cotyledons and true leaves of SER, DOB, and FLR at 4, 6, 8, and 10 dpi was determined (Table 2). No P. obducens sporangia were observed on the leaves of DOB and FLR, and a small number of sporangia could be counted on the cotyledons of FLR at 8 and 10 dpi. On the other hand, the sporangia density on the cotyledon of SER reached 3.03 × 103 sporangia cm−2 at 4 dpi, approximately three times higher than that of DOB, and at this time point, no white mildew growth could be observed on the leaves of SER yet. The sporangia density on cotyledons and leaves of SER at 10 dpi were 480 × 103 and 404 × 103 sporangia cm−2, respectively, and almost two times greater than that on the cotyledons of DOB. It showed that the susceptible levels of cotyledons and leaves of SER to P. obducens were greater than that of the cotyledons of DOB.
To assess P. obducens development on impatiens, inoculated cotyledon and true leaf segments of SER, DOB, and FLR were stained with trypan blue at 1, 2, 3, 4, 5, and 6 dpi and examined microscopically. On cotyledons of SER and DOB and true leaves of SER, similar P. obducens development was observed. Plasmopara obducens sporangia first penetrated into the adaxial leaf surface (Fig. 3B, C) and then formed vesicles, intercellular hyphae, and haustoria (Fig. 3A, E, F, G) at 1 or 2 dpi. The vesicles development in cotyledons of SER was earlier than that in cotyledons of DOB and true leaves of SER. Evident hyphae and haustoria growth were seen at 4 dpi (Fig. 3I, K) and 6 dpi (Fig. 3M–O). Monopodially branched sporangiophores first emerged from stomata at 4 dpi on cotyledons of SER and DOB, and then profuse sporangiophores and sporulation were seen on cotyledons of SER and DOB and true leaf of SER at 6 dpi (Fig. 3Q–S and Table 2). On cotyledons of DOB, apparent cell death response could be observed (Fig. 3R). On the true leaves of FLR and DOB, inoculated sporangia were observed on the adaxial leaf surface (Fig. 3D), but the development of new vesicles, hyphae, or haustoria was not seen (Fig. 3H, L, P). Therefore, the lifecycle of P. obducens did not begin in the true leaves of FLR and DOB. In the cotyledon of FLR, although the hyphae and haustoria could be observed occasionally, the extension of hyphae was greatly limited.
A–D vesicles and hyphae were observed inside the cotyledon of SER at 1 dpi (A), and sporangia attached on the surface of cotyledons of DOB (B) and true leaves of SER (C) and DOB (D). E–G Vesicles, hyphae, and haustoria were formed inside cotyledons of SER (E) and DOB (F) and true leaves of SER (G) at 2 dpi. I–K Long hyphae and haustoria were observed inside cotyledons of SER (I) and DOB (J), and true leaves of SER (K) at 4 dpi. M–O More hyphae and haustoria were seen inside cotyledons of SER (M) and DOB (N), and true leaves of SER (O) at 6 dpi. Q–S Monopodially branched sporangiophores emerged through stomata and sporangia were borne on the tips of sporangiophore branches. In Q, the sporangia were washed off during trypan blue staining. In R, the brown coloration indicated host plant cell death. H, L, P No development of vesicles, hyphae, or haustoria inside true leaves of DOB at 2, 4, and 6 dpi. CD cell death, Sp sporangia, Ve vesicle, Hy hypha, Ha haustorium, St stomata, Scale bars: 50 μm
Single-molecule real-time sequencing of transcriptomes and alternative splicing
To identify the genes controlling IDM resistance in Impatiens, we selected DOB (susceptible to IDM at the cotyledon stage, but resistant at the true leaf stage and thereafter) for full-length transcriptome characterization using the PacBio Iso-Seq protocol. In addition, SER (susceptible to IDM at the cotyledon, true leaf, and mature plant stages), FLR (resistant to IDM at the cotyledon, true leaf, and mature plant stages), and DOB were also selected for transcriptome profiling using the Illumina-based RNA-Seq methodology. To generate the full-length transcriptome of DOB, the RNA samples of cotyledon and true leaf tissues were pooled together for sequencing using the PacBio Sequel platform. A total of 716,913 raw polymerase reads (34.4 Gb) with an N50 length of 93,681 bp were generated on one SMRT cell (Supplementary Table S3). After running the bioinformatics pipeline, 464,436 circular consensus sequences (CCSs) were obtained, among which 418,648 were full-length non-chimeric (FLNC) reads containing the 5′ and 3′ primers and poly-A tails. After the clustering step, 36,954 high-quality (HQ) isoforms were generated. The minor low-quality (LQ) isoforms were excluded for further analysis. The HQ isoforms were further error-corrected using LoRDEC and trimmed RNA-Seq short reads described below. Subsequently, 36,954 error-corrected HQ isoforms were obtained with an N50 length of 3010 bp (Table 3).
To discover alternative splicing (AS) events in Impatiens, the error-corrected and non-redundant (redundancy removed using CD-HIT-EST) HQ isoforms were partitioned into transcript families by the Coding GENome reconstruction Tool (Cogent) to reconstruct full-length unique transcript models (UniTransModels). A total of 11,763 full-length UniTransModels were obtained. Based on these UniTransModels, the HQ isoforms were further collapsed using Cupcake to obtain a set of 16,752 collapsed isoforms with an N50 length of 2992 bp (Table 3). Most of these UniTransModels (8,923, 75.7%) had one isoform, while 2862 (24.3%) UniTransModels had at least two isoforms (Fig. 4A, B). Based on these UniTransModels, there were six types of AS events observed, including retained intron (RI), alternative 5′ splice-site (A5), alternative 3′ splice-site (A3), skipping exon (SE), alternative first exon (AF), and alternative last exon (AL) (Fig. 4B). Among these AS events, RI type was the most predominant (984, 64.0%), followed by A3 (286, 18.6%) and A5 (13.7%). These three types of AS events accounted for >96% of detected events. By mapping the Illumina short reads to these UniTransModels, the reliability of detected AS events was confirmed (Fig. 4C).
A Distribution of isoform numbers based on UniTransModels. B Summary of AS events identified in the full-length transcriptome using UniTransModels as the reference. RI retained intron, A5 alternative 5′ splice-site, A3 alternative 3′ splice-site, SE skipping exon, AF alternative first exon, AL alternative last exon. C Sashimi plots showing examples of the six types of AS events. Red peaks indicate overage of Illumina short reads; curved red lines with numbers represent splicing junctions supported by this number of Illumina short reads. For isoforms located at the bottom, blue blocks represent exons and lines linking blocks represent introns
The first reference transcriptome of impatiens
For each of the three cultivars (SER, FLR, and DOB), the cotyledon and true leaf tissues were also subjected to Illumina short reads sequencing (RNA-Seq). A total of 18 samples (3 cultivars × 2 tissue types × 3 replicates = 18) were sequenced. An average of 14.5 million 150-bp cleaned read pairs were obtained for each sample (Supplementary Table S4). A de novo assembly was performed for each cultivar by pooling reads of the six samples and using Trinity. A total of 118,919, 120,416, and 95,837 contigs were obtained for DOB, FLR, and SER, with an N50 length of 2112, 2164, and 2242 bp, respectively (Table 3). The three RNA-Seq assemblies were merged using TGICL with redundancy removed using CD-HIT-EST, yielding 100,049 unique transcript sequences with an N50 length of 2277 bp. The unique transcript sequences obtained from RNA-Seq were mapped to Iso-Seq isoforms. For downstream functional annotation and investigation of gene expressions, a reference transcriptome for Impatiens was constructed by combining the longest collapsed isoforms from Iso-Seq and unmapped transcript sequences from RNA-Seq. Finally, a total of 48,758 reference transcript sequences with an N50 length of 2060 bp were obtained to represent the reference transcriptome of Impatiens (Table 3). To estimate the completeness of this reference transcriptome, we compared these sequences to the BUSCO embryophyta_odb9 dataset and obtained a completeness score of 85.2%.
For functional annotation, the reference transcriptome was compared to several major public databases. The majority of the sequences (36,978; 75.8%) had hits to the non-redundant protein (NR) database, followed by Swiss-Prot (29,731; 61.0%), and non-redundant nucleotide (NT) database (22,616; 46.4%) (Supplementary Table S5). A total of 22,699 (46.6%) transcripts were annotated with gene ontology (GO) terms, with an average of four GO terms per transcript. In addition, 11,537 (23.7%) sequences were assigned with Kyoto Encyclopedia of Genes and Genomes Ontology (KO) terms. By mining the reference transcriptome in the PlantTFDB v4.0 database, a small portion of sequences (1165; 2.4%) was predicted to encode transcription factors (TFs) and assigned to TF families (Supplementary Table S6). Among the 54 TF families, the most predominant was the bHLH family (122; 10.5%), followed by bZIP (77; 6.6%), and MYB-related (62; 5.3%). By running the TransDecoder pipeline, coding regions and protein sequences were successfully predicted for a total of 34,359 (70.5%) sequences, among which 27,515 (56.4%) sequences contained a complete open reading frame (ORF). Based on the predicted proteins out of the reference transcriptome, we identified 45 NBS-containing genes and 246 leucine-rich repeat receptor-like kinase (LRR-RLK) genes (Supplementary Table S7). Among these 45 predicted NBS genes, 33 (73.3%) contained a complete ORF. These NBS genes were further classified into four types, including NBS-LRR (15), NBS (13), coiled-coil (CC)-NBS-LRR (10), and CC-NBS (7). The TIR domain was not identified in these predicted NBS genes. A phylogenetic tree was constructed based on the NBS domain sequences, which revealed two major clusters of Impatiens NBS genes (Fig. 5).
Identification of genes and R-genes potentially involved in downy mildew resistance
To identify impatiens R-genes, we first downloaded the 152 reference Pathogen Receptor Genes maintained at the PRGdb that have been cloned and well-characterized in other plant species. We also obtained 1678 proteins from NCBI and 37 Arabidopsis genes from the UniProt database based on their functionality in resistance to downy mildew. Through gene family analysis, we identified 683 impatiens genes (81 gene families) orthologous to the PRGdb reference R-genes or “downy mildew” associated genes (Supplementary Table S8). These impatiens orthologs and predicted NBS and LRR-RLK genes were prioritized for downstream evaluation of gene expressions in the five pairs of comparisons that were made possible by three cultivars (DOB, FLR, and SER) and two types of organs (cotyledon and true leaf) with different resistance or susceptibility to IDM (Fig. 6).
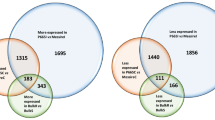
A Number of expressed transcripts (transcripts per million or TPM > 0.5) for each tissue type and each cultivar. DOB Divine Orange Bronze Leaf, SER Super Elfin Red, FLR Florific Lavender. Green circle represents cotyledon; green triangle represents true leaf. “+” indicates expressed; “−” indicates not expressed. The “No. of transcripts” (from top to bottom) corresponds to the transcripts that were only expressed in true leaves and not expressed in cotyledons for all three genotypes; transcripts that were only expressed in true leaves of Impatiens hawkeri and not expressed in cotyledons of Impatiens hawkeri, neither expressed in cotyledons nor true leaves of Impatiens walleriana; transcripts that were only expressed in cotyledons and not expressed in true leaves for all three genotypes; transcripts that were only expressed in cotyledons of Impatiens hawkeri and not expressed in true leaves of Impatiens hawkeri, neither expressed in cotyledons nor true leaves of Impatiens walleriana. B Principal component analysis of all genes using DESeq2 with normalized counts. Circles with different colors represent different groups of samples as shown in the figure. C Upregulated genes based on five downy mildew-susceptible vs. resistant sample comparisons. D Downregulated genes based on five susceptible and resistant sample comparisons
The clean reads from the 18 impatiens RNA-Seq samples were mapped to the reference transcriptome to investigate gene expression profiles of cotyledon and true leaf tissues of impatiens. By setting the transcripts per million (TPM) cutoff as 0.5 (at least one replicate) to be considered expressed, a range of 24,716–31,178 transcripts were expressed in the cotyledons and true leaves of these 18 samples (Fig. 6A and Supplementary Table S9). Apparently, a higher number of transcripts were expressed at the true leaf stage compared with the cotyledon stage for all three cultivars. Interestingly, DOB had the smallest number of expressed transcripts at the cotyledon stage but had the highest number of expressed transcripts at the true leaf stage. There were 345 transcripts only expressed at the true leaf stage for all three cultivars. A total of 1245 transcripts were only expressed at the true leaf stage for I. hawkeri samples, but not expressed in the I. walleriana sample. A much smaller number of transcripts were only expressed at the cotyledon stage. Through differential gene expression analysis using DESeq2, DOB had a much larger number of differentially expressed genes (DEGs) when transitioning from the cotyledon stage to the true leaf stage than the other two cultivars (Table 4 and Supplementary Table S10). Further principal component analysis (PCA) using DESeq2 also revealed that DOB had very different expression profiles between its cotyledons and true leaves. As shown in Fig. 6B, the expression profiles of true leaves were very similar between DOB and FLR, which belong to the same species. However, the cotyledon expression profiles of these two cultivars were separated apart. These unique features may correspond to DOB’s different responses to IDM at the cotyledon versus the true leaf stage compared with other I. hawkeri cultivars resistant to IDM at both stages. As expected, these two species were separated by the first PC, which explained 85% of the variance (PC1, 85%).
Given that DOB transitioned from IDM susceptibility (S) on cotyledons to IDM resistance (R) on true leaves, it was expected that the genes associated with resistance to IDM expressed differently in true leaves compared with cotyledons in DOB. Thus, candidate genes were first mined based on the following criteria: (1) differentially expressed (FDR < 0.05, fold change ≥ 2) for DOB cotyledon (S) vs DOB-true leaf (R); (2) within the same tissue type, for the genes upregulated in DOB-true leaf (R) compared with DOB cotyledon (S), we looked for those that were also expressed at higher levels (FDR < 0.05, fold change ≥2) in IDM-resistant cultivars than in susceptible cultivars; (3) similarly, for the genes downregulated in DOB-true leaf (R) compared with DOB cotyledon (S), we looked for those that were also expressed at lower levels (FDR < 0.05, fold change ≥2) in resistant cultivars than in susceptible cultivars (Table 4 and Supplementary Table S10). By applying these criteria, we identified 241 transcripts upregulated and 112 transcripts downregulated for all S vs R comparisons (Fig. 6C, D and Supplementary Table S11). Importantly and interestingly, three NBS genes orthologous to cloned and characterized R-genes and to those associated with DM resistance were among the 241 upregulated transcripts (Fig. 7A–C and Supplementary Table S8). These three NBS genes were expressed at significantly higher levels in all IDM-resistant samples compared with susceptible samples. For further verification, we also analyzed the gene expression data from another independent study on mature leaves (three replicates pooled for sequencing) of IDM-resistant and susceptible Impatiens cultivars41. We observed that these NBS genes were also expressed much higher in resistant Impatiens “SunPatiens® Compact Royal Magenta” (SPR) than in susceptible sample SEP. The three NBS genes were orthologous to two genes (ACY69609.1/RGC203 and ADX86902.1) in common sunflower that have been associated with resistance to Plasmopara halstedii, the causal agent of sunflower DM45,46, and were also orthologous to two genes (Rpi-blb1 and RB) conferring resistance to potato blight in Solanum bulbocastanum, a potato relative47,48. Considering this evidence, these NBS genes can be good candidates for future mining Impatiens genes conferring IDM resistance. In addition, we identified two LRR-RLK genes significantly upregulated in all IDM-resistant samples (Fig. 7D, E), which may also be candidates potentially associated with IDM resistance. The three candidate NBS genes and two candidate LRR-RLK genes were selected for qRT-PCR validation of gene expressions in I. hawkeri and I. walleriana samples. Several pairs of primers were designed for candidate gene PB.2459.1, but somehow all designed primers did not work properly for this candidate gene. The qRT-PCR results supported that the expression levels of PB.2448.1, PB. 11744.1, PB.11524.1, and CL41296Contig1 were much higher (fold change ≥2) in the resistant samples than in the susceptible samples from I. hawkeri (Fig. 8A–D). When I. walleriana samples were included in the qRT-PCR comparison, only PB.11744.1 (CC-NBS-LRR) and CL41296Contig1 (LRR-RLK) showed significantly higher expression levels in the resistant samples than in the susceptible samples. Since only a single reference gene (GAPDH) was used for normalization in these qRT-PCR analyses, caution might be warranted when comparing gene expression levels between I. hawkeri and I. walleriana.
A–C Three nucleotide-binding site (NBS)-containing genes expressed at significantly higher levels in all downy mildew-resistant samples than in susceptible samples. D, E Two leucine-rich repeat receptor-like kinase (LRR-RLK) genes expressed at significantly higher levels in all resistant samples than in susceptible samples. F Symbols and signs in A–E. DOB Divine Orange Bronze Leaf, SER Super Elfin Red, FLR Florific Lavender, SEP Super Elfin Pink, SPR SunPatiens® Compact Royal Magenta. The gene expressions in mature leaves of SEP and SPR were from a previously published, independent study41, and they were included for a comparison
The expression levels of selected candidate genes were normalized to GAPDH. A, B The relative expression of two nucleotide-binding site (NBS)-containing genes. C, D The relative expression of two leucine-rich repeat receptor-like kinase (LRR-RLK) genes. DOB Divine Orange Bronze Leaf, SER Super Elfin Red, FLR Florific Lavender
Discussion
As a worldwide challenge, DM has devastated many crops, including the ornamental crop garden impatiens, I. walleriana. Impatiens hawkeri has been reported to be generally resistant to IDM. However, to date, little research has been reported to develop disease screening or resistance phenotyping methodologies in Impatiens and to understand the IDM resistance in I. hawkeri. The lack of information on resistance phenotyping and resistance mechanism has severely hindered efforts to develop new IDM-resistant cultivars. Moreover, the interaction between P. obducens and Impatiens remains to be discovered. Therefore, to fill these gaps, we have established a rapid, efficient, and effective system to assess IDM susceptibility and resistance at very early growth stages (first and second true leaves as well as the cotyledon stage) and histologically characterized the pathogen development inside impatiens cotyledons and leaves. Using this newly developed method, we discovered that two cultivars (DL and FLR) possess strong IDM resistance starting at the earliest growth stage (the cotyledon stage) and that a number of other cultivars including DOB could make the dramatic transition within a short time and a very short distance, from susceptibility at the cotyledon stage to complete resistance at the first/second pair of true leaf stage. These cultivars and growth stages with different levels of resistance to IDM have created an excellent opportunity to investigate host–pathogen (impatiens—P. obducens) interactions and discover genes potentially involved in such a dramatic transition process. In this study, we grasped this opportunity and applied full-length transcriptome sequencing (Iso-Seq) and RNA-Seq to three cultivars (SER, DOB, and FLR) with contrasting phenotypes to IDM and made five pairs of transcriptome comparisons between IDM-resistant and susceptible cultivars and tissues. These comparisons enabled us to identify 241 transcripts upregulated in resistant cultivars and resistant tissues and three R-genes potentially involved in IDM resistance. Results and genomic resources from this study will help better understand IDM resistance in impatiens, develop molecular markers, implement genomics-assisted breeding, accelerate the development of new IDM-resistant cultivars, and provide genomic resources for cloning of IDM-resistance genes.
All I. walleriana cultivars tested in this study exhibited susceptibility to IDM throughout the entire plant developmental stages, from the cotyledon stage to mature, flowering plants. While I. hawkeri showed general resistance to IDM, many cultivars were susceptible to IDM at the cotyledon stage, indicating important influences of plant developmental stage (or tissue type) on IDM resistance or susceptibility. A similar phenomenon has been observed in certain other plant–pathogen interactions. For example, broccoli (Brassica oleracea) lines “PCB21.32” and “OL87123-2” were fully susceptible to DM (Hyaloperonospora parasitica) at the cotyledon stage but were resistant to the pathogen at 6-weeks old49. Therefore, for these plants, DM resistance cannot be predicted from cotyledon resistance. By contrast, cotyledons and true leaves in basil (Ocimum basilicum) exhibited similar DM responses, indicating that early inoculation could be used in DM resistance evaluation50. In our study, we observed different I. hawkeri cultivars had different responses to IDM at the cotyledon stage and even some of the I. hawkeri cultivars were highly susceptible to IDM at this early stage. Therefore, to prevent potential damage of IDM to these cultivars during young plant production, fungicide protection in the production facility is required at the cotyledon stage. As for breeding impatiens for IDM resistance, we recommend screening impatiens breeding populations beginning at the cotyledon stage so that newly developed resistant cultivars will have resistance through their entire plant developmental stages. Delayed IDM disease screening may result in new cultivars susceptible to this fast-acting and destructive disease at their early growth stages resulting in crop failure of large-scale young plant production. This very early stage disease screening should be invaluable to impatiens breeding: It identifies new breeding lines and ultimately new cultivars with lifetime-long resistance to IDM, and it should also greatly reduce the space, time, labor required, and costs associated with screening large numbers of impatiens breeding populations.
Hypersensitive response (HR) involves the rapid death of host cells that can limit the progress of infection. It is a plant resistance response that can be used to differentiate between resistant and susceptible plants. In different plants, varied HR symptoms have been reported when resistant hosts were infected by the DM pathogen. For instance, resistant Arabidopsis plants showed different HR symptoms against DM, such as flecking necrosis, necrosis, pitting necrosis, or trailing necrosis, depending on the strength and timing of the cell death response51. By contrast, susceptible accessions displayed heavy conidiophore sporulation without visible cell death. Compared with susceptible individuals, which did not show any visible reactions except sporulation, resistant grapevine showed HR with isolated necrosis, resulting in a significant reduction of pathogen expansion and disease symptoms52. In this study, “black spots” (and “black specks”) were observed on inoculated cotyledons of I. hawkeri. There was no simple linear correlation between disease incidence of IDM and black spot. This raised an interesting question as for what roles these black spots and specks may play in I. hawkeri’s resistance to IDM.
Although plants defend themselves against pathogens in different ways, successful host defenses disrupt the disease cycle primarily in the pre-penetration, penetration, or infection phase53. The host defense to DM in resistant plants has been studied in several plant species, such as Arabidopsis, lettuce, and grapevine52,54,55,56,57. Arabidopsis C24 appeared to develop HR upon infection with visible cell death and the elongation of hyphae branching out into the intercellular space was restricted54. In resistant transgenic lettuce, the growth process of pathogen Bremia lactucae was retarded and no sporophores were observed at any time points55. In these two examples, the lifecycle of DM pathogens in resistant plants was not completed. Whereas, in DM-resistant grapevines, P. viticola could complete its life cycle in leaf tissues, but its hyphal growth and sporangia formation were inhibited, resulting in no visible symptoms or sporulation52,56,57. The above three types of host defenses to DM all occurred in the infection phase. In comparison to above scenarios, we found that the resistance mechanism of I. hawkeri to P. obducens may be more similar to that of grapevines against P. viticola. Because in cotyledons of I. hawkeri FLR, the lifecycle of P. obducens could also complete occasionally, but hyphae growth, haustoria development, and sporulation were greatly restricted.
RNA-Seq has been a powerful approach to understanding the transcriptional regulations associated with the disease response to DM in many crops, such as grapevine57,58, spinach40, lima bean59, and pearl millet60. However, the short reads from RNA-Seq are usually insufficient to reconstruct an accurate transcriptome, especially for species without a publicly available reference genome. Our study has combined the strengths of the third-generation sequencing technology with a much longer read length and RNA-Seq in reconstructing the transcriptome for Impatiens spp. with limited genome and transcriptome resources. As revealed in this study, the N50 length of Iso-seq isoforms (2992 bp) is much longer than the final assembly from RNA-Seq (2277 bp). The full-length transcriptome not only provided full-length transcript sequences for genes but also provided insights into the AS events in Impatiens spp. In consistent with other crops such as cotton61, rice62, and Italian ryegrass63, the retained intron type contributed the majority of AS events in Impatiens. On the basis of Iso-seq isoforms and in combination with RNA-Seq assemblies, we constructed a reference transcriptome for Impatiens spp. with a BUSCO score of 85.2%. Since we only sequenced tissues collected from earlier developmental stages (cotyledon and true leaf stages), not all genes are expressed. This completeness is comparable to that of several reports in other plant species64,65.
Improving disease resistance is an important objective of impatiens breeding. Efficient screening of breeding populations to identify breeding lines with disease resistance is critical for a breeding effort toward such a goal. Development and application of molecular markers have been proposed as an effective approach to increasing the screening efficiency in impatiens disease-resistance breeding. However, few molecular markers have been reported for disease-resistance traits in impatiens, primarily due to the lack of genomic resources. The transcriptome sequences assembled in this study can play an important role in the future development of molecular markers for disease-resistance traits (and other traits) in impatiens. The 45 impatiens NBS and 246 LRR-RLK gene sequences identified in this study can be particularly valuable for this effort. It has been shown that NBS genes constitute the large family of R-genes conferring plants resistance to diverse bacterial, fungal, oomycete, and viral pathogens and nematodes, even insects in some cases66. LRR-RLK genes also play important roles in plant disease resistance, functioning as R-genes or as members of the plant defense signaling pathways. In other plants, NBS and LRR-RLK sequences often co-localize or are linked or associated with disease-resistance loci or QTLs, having allowed rapid development of new molecular markers10. Thus, these impatiens NBS and LRR-RLK sequences can serve as an excellent starting point in future efforts toward developing molecular markers for disease-resistance traits in impatiens. The full-length coding region sequences of these genes from the assembled reference impatiens genome can speed up the cloning and functional characterization of the identified impatiens disease-resistance genes.
One limitation of this study is that P. obducens-inoculated samples were not available to be included in transcriptome sequencing due to the lack of viable pathogen inoculum when this part of the study was initiated. Without P. obducens-inoculated samples in transcriptome sequencing, transcripts that were to be induced by pathogen infection could not be captured. To overcome this shortage, we made use of the unique impatiens genotype (DOB) that was discovered in this study and made comparisons between IDM-resistant and susceptible tissues and between IDM-resistant and susceptible cultivars. These comparisons enabled us to identify 241 and 112 transcripts upregulated and downregulated in IDM-resistant cultivars/tissues, respectively. These differentially expressed transcripts can be very valuable for further dissection of the interactions between impatiens and P. obducens at the molecular level. The three NBS and two LRR-RLK transcripts that were upregulated in the IDM-resistant cultivars/tissues may be of particular value because they were also expressed at higher levels in another IDM-resistant I. hawkeri cultivar in a previous paper41 and potentially involved in IDM resistance. Several approaches can be used in future experiments to test the roles of these transcripts in IDM resistance, including genetic segregation analysis, genetic mapping, gene expression analysis, genetic transformation and overexpression, and/or knockout with RNAi or gene editing67. The full-length coding region sequences of these transcripts can facilitate the initiation of all these important analyses.
For all three cultivars (DOB, FLR, and SER), a higher number of transcripts were expressed at the true leaf stage compared with the cotyledon stage, indicating that more genes are needed and induced as impatiens plants begin to grow and develop. However, it seems a smaller number of transcripts were newly induced in I. walleriana than in I. hawkeri. The I. hawkeri cultivar DOB seems a special case since the number of DEGs by comparing cotyledon and true leaf stage (5703) was at least twice of that in the other two cultivars (2163 or 1850). According to the PCA analysis, the high number of DEGs in DOB is less likely due to a distinct expression profile in true leaf, since DOB and FLR (both belonging to I. hawkeri) had very similar expression profiles in true leaf. Instead, it is more likely to be explained by the distinct expression profile of cotyledon of DOB, as DOB and FLR had relatively dissimilar expression profiles in the cotyledon. Moreover, the total number of transcripts expressed in cotyledons of DOB was only 24,716, the lowest among the three cultivars. Therefore, it is possible that some molecular or transcriptional regulations related to IDM resistance may be missing or undermined in cotyledons of DOB, but later came back to a level similar to FLR at the true leaf stage. We found these DEGs from DOB to be of particular interest since the genes associated with susceptibility (at the cotyledon stage) or resistance (at the true leaf stage) to IDM are likely among these DEGs. Subsequently, we further identified DEGs shared by all possible S vs R comparisons within the same tissue types, which could represent the transcriptional differences associated with susceptibility/resistance to IDM. The differential expression analysis combined with large-scale identification of NBS genes, LRR-RLK genes, and orthologs to public R-genes and genes associated with DM finally led to a few candidate genes, including three NBS genes.
Currently, >30 resistance genes against DM, designated as Pl genes, have been identified and extensively studied in sunflower68,69. NBS genes have played an important role. In sunflower, two types of DM resistance have been reported, including type I which restricts the pathogen growth in hypocotyls, and type II which allows the pathogen to reach hypocotyls and cotyledons45. The type II resistance (Pl14) was reported to be controlled by CC-NBS-LRR genes, while type I resistance (PlARG) likely controlled by TIR-NBS-LRR genes. Besides, the type II resistance gene (Pl14) was reported to be in close proximity to several clusters of non-TIR type NBS-LRR genes that appeared to be tandemly duplicated in the sunflower genome46. In comparison with the two types of resistance in sunflower, the resistance to IDM conferred by I. hawkeri DOB may be similar to the type II resistance. First, the cotyledons of DOB can be invaded by IDM. Second, the TIR domain was not identified in the NBS genes of impatiens, indicating most NBS genes of impatiens could be a non-TIR type. Moreover, the three NBS genes identified in this study were assigned to the same gene family with RGC203 (resistance type II) in sunflower, and two of these NBS genes are of a CC-NBS-LRR type. As a total of 20 impatiens NBS genes are assigned to this gene family, they may also belong to duplicated clusters, which needs further confirmation based on genome sequences. Future experiments can be designed to look at the temporal expressions of these candidate genes along with IDM infection and to investigate their functions. Since both susceptibility and resistance to IDM can be observed on the same plant at different growth stages, the cultivars like DOB would be an excellent plant material and model to further clarify the molecular mechanisms of IDM resistance and susceptibility in impatiens.
Conclusion
In summary, our study investigated the resistance and susceptibility of I. walleriana and I. hawkeri cultivars to P. obducens at different plant growth stages. By artificial inoculation and histological characterization of pathogen development inside inoculated tissues, we established an effective early and rapid system to screen impatiens cultivars and breeding populations for IDM resistance and to study plant–pathogen interactions. Using this system, we discovered two cultivars with strong resistance to IDM from their cotyledon stage on and additional cultivars that expressed, at different growth stages, dramatically different levels of resistance to P. obducens. We took advantage of these newly discovered disease responses and further characterized the expression profiles of cotyledons and true leaves of Impatiens. Our study has provided a comprehensive data source for mining disease-resistance genes in Impatiens, including transcriptome-wide identified NBS genes, LRR-RLK genes, genes orthologous to public R-genes and downy mildew associated genes, and DEGs differentially regulated between resistant and susceptible cultivars and tissues. Our results have laid a solid foundation for further research to understand and improve DM resistance in impatiens and have good potential to be applied to other crops.
Materials and methods
Impatiens walleriana and I. hawkeri cultivars and seedlings
Sixteen cultivars of I. walleriana from Accent Premium, Xtreme, Super Elfin, and Balance series and 16 I. hawkeri from Florific and Divine series (Table 1 and Supplementary Table S1) were evaluated for their response to P. obducens infection at the cotyledon, first/second pair of true leaf, and mature plant stages. Seeds of these 32 cultivars were sown on 20-rowed germination trays (model P-SEED20; Landmark Plastic Co., Orlando, FL) filled with Fafard germination Mix (Conrad Fafard, Inc., Agawam, MA). The trays were covered with plastic lids to keep moisture in a growth room at temperatures between 22 and 25 °C and a photoperiod of 16 h light/8 h dark. Seedlings with cotyledons (about two weeks old for I. walleriana and three weeks old for I. hawkeri) were transferred, one plant per cell, into 128-cell trays (model TR128D; Speedling Inc., Sun City, FL) filled with the commercial potting mix Fafard 3B mix (Conrad Fafard, Inc.). Seedlings were grown in the DM-free greenhouse with the temperature controlled between 25 and 30 °C. A liquid fertilizer containing 20% (w/w) nitrogen, 20% (w/w) phosphate (P2O5), and 20% (w/w) potassium (K2O) (Southern Agricultural Insecticides Inc., Palmetto, FL) was applied to the seedlings at 75 ppm twice a week following the irrigation program. All seedlings used in different experiments were grown using this method and all experiments were conducted at the University of Florida’s Gulf Coast Research and Education Center (UF/GCREC) (lat. 27°45’36” N, long. 82°13’45” W; AHS Heat Zone 10; USD Cold Hardiness Zone 9 A) in Wimauma, FL, USA.
Plant growth and field disease evaluation
On April 11, 2014 (47 days after seeds were sown), seedlings with four pairs of true leaves were transplanted into 72-cell trays (model TR72D; Speedling Inc., Sun City, FL) filled with the commercial potting mix Fafard 3B mix and kept in the greenhouse. The same liquid fertilizer was applied to the seedlings at 75 ppm once a day following the irrigation program. On June, 5 2014 (0 DAP), plants were transplanted on the 20-cm-high, 81-cm-wide raised ground beds of EauGallie fine sand covered with white-on-black plastic mulch in the experimental field of UF/GCREC. The overhead shade cloth was set up over the beds to create a partially shady environment (~40%). All cultivars were planted following a randomized complete block design with three blocks. For each cultivar, two biological replications in each block were grown 112.5 cm apart from each other. Drip irrigation with regular fertilizer and insecticide programs were followed. Plants were checked visually every 2 days for white sporulation on the abaxial side of leaves as an indication of IDM. Diseased leaves were sampled and observed under a bright-field microscope (BH-2) to confirm the pathogen identity.
In vivo preservation of DM pathogen
Plasmopora obducens sporangia were obtained from I. walleriana Accent Premium Rose (APR) during a field trial in March 2015 and then used to inoculate susceptible I. walleriana APR stock plant maintained in an isolated growth room. First, identification of P. obducens causing DM was achieved by symptoms of plants, and the morphology of sporangiophores and sporangia described by Palmateer et al.24. A sporangia solution (1 × 105 sporangia ml−1) was prepared as described by Pyne et al.50. Fresh sporulating leaves of APR were dipped into distilled water and gently agitated for 5 min. The P. obducens sporangia suspension was filtered through a 40-μm nylon mesh cell strainer (Thermo Fisher Scientific, Bridgewater, NJ) and then centrifuged at 3000×g for 10 min. This mesh cell strainer was used to remove debris and produce a cleaner sporangia suspension. Since P. obducens sporangia were ovoid and 12.7–25.0 × 10.0–17.7 µm in dimension24,29, they were expected to pass through the strainer easily. The supernatant was discarded, leaving the pellet re-suspended in 10 ml of distilled water. The sporangia density in the suspension was adjusted to a final density of 1 × 105 sporangia ml−1 using a Reichert Bright-Line hemocytometer (Hausser Scientific, Horsham, PA) and a BH-2 microscope (Olympus America Inc., Melville, NY). The prepared sporangia suspension was finely sprayed onto the adaxial leaf surface of I. walleriana APR plants (60-days old). The inoculated plants were kept in closed plastic bags on a metal bench in the growth room with the air temperature maintained at 21 ± 1°C, light intensity of 160 µmol m–2 s–1, and 16-h light/8-h dark. The humidity inside the plastic bags was 100%, measured with hygrometers. After 7 days post inoculation (dpi), white downy growth was visualized on the abaxial leaf surface. Disease symptoms and the morphology of the sporangiophores and sporangia were compared to the control plants to verify the pathogen.
Inoculating I. walleriana and I. hawkeri seedlings at three growth stages
Seedlings of 16 I. walleriana and 16 I. hawkeri cultivars were individually inoculated with P. obducens sporangia at the cotyledon stage or the first/second pair of true leaf stage in 128-cell trays. One droplet (~20 µL per droplet) of sporangia suspension (1 × 105 sporangia mL−1) was added to the adaxial and abaxial sides of each cotyledon, respectively. To inoculate the first/second pair of true leaves, five droplets were applied onto the adaxial and abaxial sides of each leaf, respectively. Inoculated seedlings were immediately enclosed inside a polythene bag for 20 days. In the control treatment (non-inoculated), the cotyledons and first/second pair of true leaves were mock-inoculated with the same numbers of droplets of distilled water, and these seedlings were kept in a separate growth room with the same growing conditions. Inoculated seedlings were evaluated for IDM disease symptoms at 10 and 20 dpi. The disease incidence index was defined as mean ratings for downy mildew incidence using a binary scale, in which 0 equaled no visible sporulation and 1 equaled visible sporulation on the abaxial side of the cotyledon. A randomized complete block design with three replicates was used as the experimental design. For each replicate, 12 or 16 cotyledons or leaves were sampled per cultivar. Data analysis was performed in SAS version 8.1 (SAS Institute Inc., Cary, NC).
Observation of pathogen development in inoculated cotyledons and true leaves
Based on in vivo inoculation results, I. walleriana SER, I. hawkeri DOB, and FLR were selected for microscopic observation. Infected cotyledon and true leaf segments (~5 × 10 mm) were collected, washed with autoclaved distilled water three times, and placed on 1% autoclaved water agar in plastic disposable Petri dishes (9.5 cm in diameter; 20 mL per dish). Cotyledon and leaf segments were inoculated with one 10-µL droplet at 1 × 105 sporangia mL−1 on the adaxial surface. The inoculated cotyledon and true leaf segments were incubated for 24 h under the above-described conditions. Thereafter, the sporangia suspension droplets were blotted with autoclaved filter papers. The cotyledon and true leaf segments were kept on the water agar with the abaxial surface up to observe disease symptoms.
The development of P. obducens in impatiens cotyledons or true leaves was examined by microscopic observation of trypan blue-stained impatiens tissues. Five inoculated cotyledon or leaf segments per treatment were removed from the Petri dishes at 1, 2, 3, 4, 5, and 6 dpi and fixed by soaking them in 5 mL of the clearing solution A (acetic acid:ethanol = 1:3, v/v) in a 50-mL tube (one or two segments per tube). Tubes were shaken at a low speed (80 rpm) overnight. Subsequently, the clearing solution A was removed and replaced with 5 mL of the clearing solution B (acetic acid:ethanol:glycerol = 1:5:1, v/v/v). The tissue samples were shaken for at least 3 h and then treated with 5 mL of 0.01 % trypan blue (Sigma-Aldrich) staining solution (trypan blue:lactic acid:phenol:distilled water = 0.003:1:1:1, w/v/v/v). Impatiens tissue samples were stained overnight on a shaker at a low speed (80 rpm). Stained cotyledon or leaf tissues were rinsed with a small amount of autoclaved 60% glycerol to remove the staining solution, immersed in 5 mL of autoclaved 60% glycerol, and shaken at 80 rpm for at least 2 h. Finally, the well-stained impatiens tissue samples were placed on a clean glass slide in a drop of 60% glycerol, covered with a coverslip, and observed under a microscope (BX41) equipped with an Olympus Q-color 5 camera (Olympus America Inc., Melville, NY).
Determination of sporangia densities on inoculated cotyledons and true leaves
Small pieces (5 × 5 mm) of tissue from inoculated cotyledon and leaf segments were cut and immersed in 200 µL distilled water amended with Tween 20 (0.05%; Sigma-Aldrich, St. Louis, MO) in a 1.5-mL microcentrifuge tube. The tubes were vortexed on a mini-shaker (Vortex-Genie; Fisher Scientific, Waltham, MA) for 5–10 s to dislodge the sporangia from cotyledon or leaf surfaces. Sporangia in the suspension were counted using a hemocytometer under a bright-field microscope (BH-2). Sporangia counts were converted into sporangia densities (total number of sporangia/area of the cotyledon or leaf segment sampled). Sporangia counting was performed every two days from 4 dpi to 10 dpi. For each time point, eight pieces of cotyledon or leaf segments were sampled. This experiment was repeated three times.
Library preparation and sequencing
To investigate the normal transcriptome profiles of IDM-resistant and susceptible cultivars at cotyledon and true leaf stages, three representative cultivars were selected, including DOB (susceptible to IDM at the cotyledon stage; resistant to IDM at the true leaf stage and thereafter), SER (susceptible to IDM at all stages), and FLR (resistant to IDM at all stages). Seeds were planted on a commercial potting mix in containers and germinated in a greenhouse facility at UF/GCREC, USA. Cotyledons and first true leaves were collected at the cotyledon stage and true leaf stage, respectively, without P. obducens inoculation. For each tissue type/cultivar, samples were collected from three biological replicates. Collected samples were immediately frozen in liquid nitrogen for RNA extraction. RNA samples were extracted using RNeasy Plus Mini kit (Qiagen, CA, USA). RNA quality and quantity were evaluated using Qubit fluorometer 2.0 (Thermo Fisher Scientific, Waltham, USA) and Agilent 2100 Bioanalyzer (Agilent Technologies, CA, USA), respectively. The RNA samples of cotyledons and true leaves of DOB were pooled in equal amounts for PacBio Iso-seq. The SMARTer PCR cDNA Synthesis Kit (Clontech, CA, USA) was used for full-length cDNA synthesis. Two size bins (<3 Kb and >3 Kb) were used for cDNA fraction and Iso-Seq library construction, which was sequenced on one SMRT cell of the PacBio Sequel system (PacBio, CA, USA) at the Interdisciplinary Center for Biotechnology Research, University of Florida, Gainesville, FL, USA. In addition, the RNA samples from the above three cultivars were sent to the University of California Davis Genome Center for Illumina HiSeq4000 and NovaSeq6000 sequencing (150 bp paired-end reads).
Iso-Seq and RNA-Seq data analysis
The Iso-Seq raw data were processed following the PacBio Iso-Seq pipeline using SMRT Link v8.0 and Iso-Seq3 (https://github.com/PacificBiosciences/IsoSeq_SA3nUP). Only high-quality (HQ) consensus sequences were used for further analysis. The trimmed Illumina short reads below were used to correct errors in the HQ consensus sequences using LoRDEC70. The redundancy was removed using CD-HIT-EST71 (-c 0.99 -n 10 -T 0 -M 0 -r 1). Cogent v2.1 was used to reconstruct the unique transcript models (UniTransModels) (https://github.com/Magdoll/Cogent). The error-corrected and non-redundant HQ consensus sequences were mapped to the UniTransModels using GMAP and further collapsed using Cupcake (https://github.com/Magdoll/cDNA_Cupcake). The alternative splicing (AS) events were identified using SUPPA72 with default settings. For visualization of AS events, sashimi plots were generated using the Integrative Genomics Viewer (IGV). The bam file was obtained by aligning the Illumina short reads of DOB to the UniTransModels using Tophat273.
The raw Illumina reads (HiSeq and NovaSeq) were trimmed using Trimmomatic74. The trimmed reads belonging to the same cultivar were pooled for a de novo assembly using Trinity75, respectively (--min_kmer_cov 2). The three resulting assemblies were merged using TGICL v2.1 with default options76. Redundancy was removed using CD-HIT-EST (-c 0.95 -n 9 -T 0 -M 0 -r 1). To construct a reference transcriptome for downstream annotation and gene expression analyses, the final transcripts from RNA-Seq were mapped to the Iso-seq isoforms using BWA-mem77. The longest isoforms from Iso-seq and unmapped transcripts from RNA-Seq assembly were combined to represent the reference transcriptome of Impatiens spp. To assess the completeness of the reference transcriptome, it was compared to the BUSCO OrthoDB9 embryophyta dataset.
Functional annotation and prediction of coding sequences
The reference transcriptome was compared to the non-redundant protein (nr), non-redundant nucleotide (nt) databases from NCBI (https://www.ncbi.nlm.nih.gov/), Swiss-Prot database (https://www.uniprot.org/), and Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kaas-bin/kaas_main) using Blast (E-value ≤1e-05). Gene ontology (GO) terms were assigned using Blast2Go78 (-v -annot -dat -img -ips ipsr -annex -goslim). The Plant Transcription Factor Database (PlantTFDB) v4.0 (http://planttfdb.cbi.pku.edu.cn/prediction.php) was used to predict transcription factors (TFs). The coding sequences (CDS) and protein sequences were predicted following the TransDecoder pipeline (https://github.com/TransDecoder/TransDecoder) integrating the Blast (Swiss-Prot) and Pfam search results.
The NBS-containing genes were predicted by searching (hmmsearch) the predicted protein sequences using the hidden Markov model (HMM) profile of the NBS (PF00931)79 under E-value 1 × 10−4. PfamScan and NCBI Conserved Domain Search were used for confirmation of the NBS domain. The classification of NBS genes based on TIR, LRR, and CC domains was performed using NCBI Conserved Domains tool and Marcoil80 (probability > 90%). The NBS domain sequences were retrieved to construct a phylogenetic tree using RAxML under ‘PROTGAMMAJTTF’ model with 1000 bootstraps81. The identification of LRR-RLK genes followed the same method described previously82.
Comprehensive search for downy mildew associated genes and gene family analysis
To collect publicly available plant proteins associated with DM, the keyword “downy mildew” was first searched at NCBI and 1678 proteins were obtained. The keyword was also searched at UniProt and 37 proteins from Arabidopsis were obtained. In addition, the 152 reference Pathogen Receptor Genes maintained at PRGdb (http://prgdb.org/prgdb/) were also included. To identify Impatiens orthologs, gene family analysis was performed for the above-collected proteins and predicted proteins from Impatiens using All-Against-All Blast (E-value 1 × 10−5) and OrthoMCL83.
Differential expression analysis
The clean reads for each replicate were aligned to the reference transcriptome using BWA-mem. Only uniquely mapped reads were considered for further analysis. Read counts were obtained using HTSeq84. Differentially expressed genes (DEGs) were identified using DESeq285 under the cutoff of false discovery rate (FDR) < 0.05 and fold change ≥2. The transcripts per million (TPM) values were calculated using TPMCalculator86.
qRT-PCR validation
Two NBS genes and two LRR-RLK genes were selected for validation of gene expression in I. hawkeri samples using qRT-PCR. Primers were designed using BatchPrimer3 v1.0 (http://probes.pw.usda.gov/batchprimer3/). The RNA samples of cotyledon and true leaf tissues for DOB and FLR were used for cDNA synthesis with the SuperScript® III First-Strand Synthesis System for RT-PCR kit (Invitrogen, CA, USA). qRT-PCR was carried out with three biological replicates and each containing two technical replicates for each tissue type using the Power SYBR® Green PCR Master Mix kit (Applied Biosystems, USA). The cDNA levels of selected genes were normalized to the reference gene GAPDH.
Data availability
The sequencing data and transcript assemblies generated from this study have been deposited at GenBank under SRA accession number PRJNA700505.
References
Thines, M., Voglmayr, H. & Göker, M. in Oomycete Genetics and Genomics: Diversity, Interactions, and Research Tools. (eds Lamour, K. & Kamoun, S.) 47–75 (John Wiley & Sons, 2009).
Delmas, C. E. et al. Adaptation of a plant pathogen to partial host resistance: selection for greater aggressiveness in grapevine downy mildew. Evol. Appl. 9, 709–725 (2016).
Zhang, N., Lindhout, P., Niks, R. & Jeuken, M. Genetic dissection of Lactuca saligna nonhost resistance to downy mildew at various lettuce developmental stages. Plant Pathol. 58, 923–932 (2009).
Meena, P., Thomas, L. & Singh, D. Assessment of yield losses in Brassica juncea due to downy mildew (Hyaloperonospora brassicae). J. Oilseed Brassica 1, 73–77 (2016).
Chelpuri, D. et al. Mapping quantitative trait loci (QTLs) associated with resistance to major pathotype-isolates of pearl millet downy mildew pathogen. Eur. J. Plant Pathol. 154, 983–994 (2019).
Lebeda, A., Křístková, E., Sedláková, B. & Widrlechner, M. P. Recent advances in cucurbit downy mildew research and their contribution to the development of integrated protection of cucurbits. Ecol. Evol. Org. Biol. Proc. 3. https://lib.dr.iastate.edu/eeob_ag_conf/3 (2019).
Delmas, C. E. et al. Soft selective sweeps in fungicide resistance evolution: recurrent mutations without fitness costs in grapevine downy mildew. Mol. Ecol. 26, 1936–1951 (2017).
Callan, B. & Carris, L. in Biodiversity of Fungi: Inventory and Monitoring Methods. (eds Mueller, G. M., Bills, G. F. & Foster, M. S.) 105–126 (Elsevier Academic Press, 2004).
Pecrix, Y., Penouilh-Suzette, C., Muños, S., Vear, F. & Godiard, L. Ten broad spectrum resistances to downy mildew physically mapped on the sunflower genome. Front. Plant Sci. 9, 1780 (2018).
Parra, L. et al. Rationalization of genes for resistance to Bremia lactucae in lettuce. Euphytica 210, 309–326 (2016).
Vezzulli, S. et al. The Rpv3-3 haplotype and stilbenoid induction mediate downy mildew resistance in a grapevine interspecific population. Front. Plant Sci. 10, 234 (2019).
Radwan, O., Bouzidi, M. F., Nicolas, P. & Mouzeyar, S. Development of PCR markers for the Pl5/Pl8 locus for resistance to Plasmopara halstedii in sunflower, Helianthus annuus L. from complete CC-NBS-LRR sequences. Theor. Appl. Genet. 109, 176–185 (2004).
Michelmore, R. & Wong, J. Classical and molecular genetics of Bremia lactucae, cause of lettuce downy mildew. Eur. J. Plant Pathol. 122, 19 (2008).
Hermanns, M., Slusarenko, A. J. & Schlaich, N. L. Organ-specificity in a plant disease is determined independently of R gene signaling. Mol. Plant-Microbe Interact 16, 752–759 (2003).
She, H. et al. Fine mapping and candidate gene screening of the downy mildew resistance gene RPF1 in Spinach. Theor. Appl. Genet. 131, 2529–2541 (2018).
Morgan, R. J. (ed.) Impatiens: the Vibrant World of Busy Lizzies, Balsams, and Touch-me-nots (Timber Press, 2007).
Lim, T. (ed.) Edible Medicinal and Non-medicinal Plants (Springer, 2014).
Uchneat, M. S. (ed.) Flower Breeding and Genetics (Springer, 2007).
US Department of Agriculture, National Agricultural Statistics Service. https://www.nass.usda.gov (2019).
Wang, W., He, Y., Cao, Z. & Deng, Z. Induction of tetraploids in impatiens (Impatiens walleriana) and characterization of their changes in morphology and resistance to downy mildew. HortScience 53, 925–931 (2018).
Vajna, L. First report of Plasmopara obducens on impatiens (Impatiens walleriana) in Hungary. New Dis. Rep 24, 2044–0588 (2011).
Cunnington, J., Aldaoud, R., Loh, M., Washington, W. & Irvine, G. First record of Plasmopara obducens (downy mildew) on impatiens in Australia. Plant Pathol. 57, 371 (2008).
Crouch, J., Ko, M. & McKemy, J. First report of impatiens downy mildew outbreaks caused by Plasmopara obducens throughout the Hawai’ian islands. Plant Dis. 98, 696–696 (2014).
Palmateer, A., Lopez, P., Seijo, T. & Peres, N. Severe outbreak of downy mildew caused by Plasmopara obducens on Impatiens walleriana in Florida. Plant Dis. 97, 687–687 (2013).
US Department of Agriculture, National Agricultural Statistics Service. https://www.nass.usda.gov (2016).
Conner, K., Olive, J., Hagan, A., Zhang, L. & Bloodworth, M. First report of impatiens downy mildew caused by Plasmopara obducens in Alabama. Plant Dis. 98, 1006–1006 (2014).
Warfield, C. Y. Downy mildew of impatiens. GrowerTalks 75, 78–86 (2012).
Hansen, M. A., Bush, E. A., Latimer, J. G. & Hong, C. Impatiens Downy Mildew (Virginia Coop Extension Virgia State Univ., 2013).
Satou, M., Sugawara, K., Nagashima, S., Tsukamoto, T. & Matsushita, Y. Downy mildew of busy lizzie caused by Plasmopara obducens in Japan. J. Gen. Plant Pathol. 79, 205–208 (2013).
Jones, D. & O’Neill, T. Impatiens downy mildew. East Malling, UK: Horticultural Development Council FactSheet 5, 8 (2004).
Choi, Y. J., Han, J. G., Park, M. J. & Shin, H. D. Downy mildew of Impatiens balsamina and I. walleriana in Korea. Plant Pathol. J. 25, 433–433 (2009).
Keach, J. E. & Bridgen, M. Towards improvement of Impatiens. Acta Hortic. 1140, 317–325 (2016).
Eskandari, F. & Shishkoff, N. Systemic infection of Impatiens balsamina through inoculation of roots with vegetative sporangia of the Impatiens downy mildew (Plasmopara obducens). Phytopathol. 107, S4.3 (2017).
Suarez, S. & Palmateer, A. Overview of impatiens downy mildew in Florida. Phytopathol. 104, 115 (2014).
Suarez, S., Lopez, P., Chase, A. & Palmateer, A. Preventative fungicide applications in production and their impact on residual efficacy against impatiens downy mildew in the landscape. Phytopathol. 106, 68 (2016).
Gessler, C., Pertot, I. & Perazzolli, M. Plasmopara viticola: a review of knowledge on downy mildew of grapevine and effective disease management. Phytopathol. Mediterr. 50, 3–44 (2011).
Stassen, J. H. et al. Effector identification in the lettuce downy mildew Bremia lactucae by massively parallel transcriptome sequencing. Mol. Plant Pathol. 13, 719–731 (2012).
Li, X. et al. Comparative transcriptome analysis reveals defense-related genes and pathways against downy mildew in Vitis amurensis grapevine. Plant Physiol. Biochem. 95, 1–14 (2015).
Mestre, P. et al. Comparative analysis of expressed CRN and RXLR effectors from two Plasmopara species causing grapevine and sunflower downy mildew. Plant Pathol. 65, 767–781 (2016).
Kandel, S. L. et al. Transcriptional analyses of differential cultivars during resistant and susceptible interactions with Peronospora effusa, the causal agent of spinach downy mildew. Sci. Rep. 10, 1–13 (2020).
Bhattarai, K., Wang, W., Cao, Z. & Deng, Z. Comparative analysis of impatiens leaf transcriptomes reveal candidate genes for resistance to downy mildew caused by Plasmopara obducens. Int. J. Mol. Sci. 19, 2057 (2018).
Suarez, S., Naveed, Z. A. & Ali, G. Transcriptional profiling of Impatiens walleriana genes through different stages of downy mildew infection reveals novel genes involved in disease susceptibility. Preprint at https://www.biorxiv.org/content/10.1101/622480v2 (2019).
Rhoads, A. & Au, K. F. PacBio sequencing and its applications. Genomics Proteomics Bioinformatics 13, 278–289 (2015).
Florida Automated Weather Network. https://fawn.ifas.ufl.edu (2015).
Radwan, O., Bouzidi, M. F. & Mouzeyar, S. Molecular characterization of two types of resistance in sunflower to Plasmopara halstedii, the causal agent of downy mildew. Phytopathol. 101, 970–979 (2011).
Bachlava, E. et al. Downy mildew (Pl8 and Pl14) and rust (RAdv) resistance genes reside in close proximity to tandemly duplicated clusters of non-TIR-like NBS-LRR-encoding genes on sunflower chromosomes 1 and 13. Theor. Appl. Genet. 122, 1211–1221 (2011).
Song, J. et al. Gene RB cloned from Solanum bulbocastanum confers broad spectrum resistance to potato late blight. Proc. Natl Acad. Sci. USA 100, 9128–9133 (2003).
Van Der Vossen, E. et al. An ancient R gene from the wild potato species Solanum bulbocastanum confers broad-spectrum resistance to Phytophthora infestans in cultivated potato and tomato. Plant J. 36, 867–882 (2003).
Coelho, P. S., Valério, L. & Monteiro, A. A. Leaf position, leaf age and plant age affect the expression of downy mildew resistance in Brassica oleracea. Eur. J. Plant Pathol. 125, 179–188 (2009).
Pyne, R. M., Koroch, A. R., Wyenandt, C. A. & Simon, J. E. A rapid screening approach to identify resistance to basil downy mildew (Peronospora belbahrii). HortScience 49, 1041–1045 (2014).
Coates, M. E. & Beynon, J. L. Hyaloperonospora arabidopsidis as a pathogen model. Ann. Rev. Phytopathol. 48, 329–345 (2010).
Bellin, D. et al. Resistance to Plasmopara viticola in grapevine ‘Bianca’is controlled by a major dominant gene causing localised necrosis at the infection site. Theor. Appl. Genet. 120, 163–176 (2009).
Trigiano, R. N. in Plant Pathology Concepts and Laboratory Exercises (eds Ownley, B. H. & Trigiano, R. N.) 349–356 (CRC Press, 2007).
Lapin, D., Meyer, R. C., Takahashi, H., Bechtold, U. & Van den Ackerveken, G. Broad-spectrum resistance of Arabidopsis C24 to downy mildew is mediated by different combinations of isolate-specific loci. New Phytol. 196, 1171–1181 (2012).
Govindarajulu, M., Epstein, L., Wroblewski, T. & Michelmore, R. W. Host‐induced gene silencing inhibits the biotrophic pathogen causing downy mildew of lettuce. Plant Biotechnol. J. 13, 875–883 (2015).
Diez-Navajas, A., Wiedemann-Merdinoglu, S., Greif, C. & Merdinoglu, D. Nonhost versus host resistance to the grapevine downy mildew, Plasmopara viticola, studied at the tissue level. Phytopathol. 98, 776–780 (2008).
Yin, X. et al. Pathogen development and host responses to Plasmopara viticola in resistant and susceptible grapevines: an ultrastructural study. Hortic. Res. 4, 17033 (2017).
Perazzolli, M. et al. Downy mildew resistance induced by Trichoderma harzianum T39 in susceptible grapevines partially mimics transcriptional changes of resistant genotypes. BMC Genomics 13, 660 (2012).
Kunjeti, S. G. et al. RNA‐Seq reveals infection‐related global gene changes in Phytophthora phaseoli, the causal agent of lima bean downy mildew. Mol. Plant Pathol. 13, 454–466 (2012).
Kulkarni, K. S. et al. De novo transcriptome sequencing to dissect candidate genes associated with pearl millet-downy mildew (Sclerospora graminicola Sacc.) interaction. Front. Plant Sci. 7, 847 (2016).
Wang, M. et al. A global survey of alternative splicing in allopolyploid cotton: landscape, complexity and regulation. New Phytol. 217, 163–178 (2018).
Zhang, G. et al. PacBio full‐length cDNA sequencing integrated with RNA‐seq reads drastically improves the discovery of splicing transcripts in rice. Plant J. 97, 296–305 (2019).
Hu, Z. et al. Full-length transcriptome assembly of Italian ryegrass root integrated with RNA-Seq to identify genes in response to plant cadmium stress. Int. J. Mol. Sci. 21, 1067 (2020).
Feng, S., Xu, M., Liu, F., Cui, C. & Zhou, B. Reconstruction of the full-length transcriptome atlas using PacBio Iso-Seq provides insight into the alternative splicing in Gossypium australe. BMC Plant Biol. 19, 1–16 (2019).
Kuang, X., Sun, S., Wei, J., Li, Y. & Sun, C. Iso-Seq analysis of the Taxus cuspidata transcriptome reveals the complexity of Taxol biosynthesis. BMC Plant Biol. 19, 210 (2019).
Tan, X. et al. Global expression analysis of nucleotide binding site-leucine rich repeat-encoding and related genes in Arabidopsis. BMC Plant Biol. 7, 1–20 (2007).
Li, M. Y. et al. CRISPR/Cas9-mediated VvPR4b editing decreases downy mildew resistance in grapevine (Vitis vinifera L.). Hortic. Res. 7, 149 (2020).
Qi, L., Ma, G., Li, X. & Seiler, G. Diversification of the downy mildew resistance gene pool by introgression of a new gene, Pl35, from wild Helianthus argophyllus into oilseed and confection sunflowers (Helianthus annuus L.). Theor. Appl. Genet. 132, 2553–2565 (2019).
Ma, G. et al. Molecular dissection of resistance gene cluster and candidate gene identification of Pl17 and Pl19 in sunflower by whole-genome resequencing. Sci. Rep. 9, 1–10 (2019).
Salmela, L. & Rivals, E. LoRDEC: accurate and efficient long read error correction. Bioinformatics 30, 3506–3514 (2014).
Li, W. & Godzik, A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006).
Alamancos, G. P., Pagès, A., Trincado, J. L., Bellora, N. & Eyras, E. Leveraging transcript quantification for fast computation of alternative splicing profiles. RNA 21, 1521–1531 (2015).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011).
Pertea, G. et al. TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics 19, 651–652 (2003).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Conesa, A. et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676 (2005).
Finn, R. D., Clements, J. & Eddy, S. R. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37 (2011).
Delorenzi, M., Speed, T. & An, H. M. M. model for coiled-coil domains and a comparison with PSSM-based predictions. Bioinformatics 18, 617–625 (2002).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Magalhães, D. M. et al. LRR-RLK family from two Citrus species: genome-wide identification and evolutionary aspects. BMC Genomics 17, 623 (2016).
Fischer, S. et al. Using OrthoMCL to assign proteins to OrthoMCL‐DB groups or to cluster proteomes into new ortholog groups. Curr. Protoc. Bioinformatics 35, 6.12.1–6.12.19 (2011).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Vera Alvarez, R., Pongor, L. S., Mariño-Ramírez, L. & Landsman, D. TPMCalculator: one-step software to quantify mRNA abundance of genomic features. Bioinformatics 35, 1960–1962 (2019).
Acknowledgements
This research was supported by grants from the Florida Department of Agriculture and Consumer Service (FDACS) Florida Specialty Crop Block Grant Program (FDACS contract # 020705 to A.P. and Z.D.), the Fred C. Gloeckner Foundation, Inc. (Z.D.), and the USDA/NIFA hatch projects (Project # FLA-GCR-005065 and FLA-GCC-005507) (Z.D.). We thank Gail Bowman and Joyce Jones for their assistance in caring for impatiens seedlings. The authors thank PanAmerica Seed, Syngenta, and Hemgenetics for providing seeds of Impatiens walleriana and I. hawkeri cultivars, Dr. David Moraga at the University of Florida (UF) Interdisciplinary Center for Biotechnology Research for performing PacBio sequencing, the University of California Davis Genome Center for performing Illumina-based RNA sequencing, and the UF Research Computing for providing the HiPerGator for bioinformatic analysis.
Author information
Authors and Affiliations
Contributions
Z.D. designed and supervised the project. A.J.P. secured funding and identified and confirmed the pathogen. Y.H. and W.W. performed impatiens downy mildew resistance phenotyping and evaluated sporangia densities and plant–pathogen interactions. Z.P. and S.P. prepared materials for transcriptome sequencing. Z.P. and Q.Y. performed bioinformatics data analysis. Z.P., Y.H., and K.B. wrote the original manuscript draft. Z.P. and Z.D. reviewed and revised the manuscript. All authors reviewed and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Peng, Z., He, Y., Parajuli, S. et al. Integration of early disease-resistance phenotyping, histological characterization, and transcriptome sequencing reveals insights into downy mildew resistance in impatiens. Hortic Res 8, 108 (2021). https://doi.org/10.1038/s41438-021-00543-w
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41438-021-00543-w
This article is cited by
-
Adaptation mechanism of three Impatiens species to different habitats based on stem morphology, lignin and MYB4 gene
BMC Plant Biology (2024)
-
QTL Mapping and Genetic Map for the Ornamental Sunflower in China
Plant Molecular Biology Reporter (2024)