Abstract
The cwp (cuticular water permeability) gene controls the development of cuticular microfissuring and subsequent fruit dehydration in tomato. The gene underwent silencing in the evolution of the fleshy cultivated tomato but is expressed in the primitive wild tomato relatives. The introgression of the expressed allele from the wild S. habrochaites (cwph) into the cultivated tomato (Solanum lycopersicum) leads to the phenotype of fruit water loss during and following ripening. In this report, we show that low temperature impacts on the severity of the cuticular microfissure phenotype via a combination of effects on both expression and alternative splicing of cwph. The cwp gene, comprising four exons and three introns, undergoes post-transcriptional alternative splicing processes, leading to seven alternative transcripts that differ in reading-frame lengths. Transgenic plants expressing each of the alternative transcripts identified the longest reading frame (VAR1) as the functional splice variant. Low temperature led to a strong upregulation of cwph expression, compounded by an increase in the relative proportion of the functional VAR1 transcript, leading to increased severity of microfissuring of the cuticle. In summary, we demonstrate the molecular mechanism behind the horticultural phenomenon of the low-temperature effect on cuticular microfissures in the dehydrating tomato.
Similar content being viewed by others
Introduction
The aerial organs of all plants are covered by a thin layer composed of a polyester matrix of cutin and organic waxes, the cuticle, in concert with the epidermal cell walls, which protects the inner plant organs against biotic and abiotic stresses, especially loss of water1,2. The cultivated tomato fruit is covered by a relatively thick cuticle and lacks stomata, making the fleshy fruit cuticle a continuous tissue relatively impervious to water loss even following harvest3,4,5. The fruits of the various wild tomato relatives exhibit a wide range in size, color, and texture6, including diverse cuticular architecture and chemical composition, but the relatively thick cuticle is particularly characteristic of the cultivated S. lycopersicon7.
Fleshy fruit may develop fissures in the cuticle, of varying sizes and spatial organization, and this is true as well for tomato8. Cuticular microfissures are generally considered an undesirable phenotype and can lead to reduction in fruit quality and yield loss8,9. In tomato fruit, they are controlled both by genetic factors, including genes that regulate the biosynthesis of the cuticle and cell wall components10,11,12,13,14,15 and by environmental factors, including irrigation, temperature, humidity, and intense exposure to radiation8,16,17. The resultant chemical components and the related physical and chemical structure impacts on the biomechanical characteristics of the fruit cuticle tissue18,19,20,21.
Previously, we identified the gene cwp (cuticular water permeability) whose expression underwent silencing during the evolution and domestication of the cultivated fleshy fruit tomato3. While the allele from the cultivated S. lycopersicon (cwpe, from the former nomenclature Lycoperiscum esculentum) is not expressed, the alleles of the primitive green-fruited species are expressed during early fruit development. Significantly, when the cwp gene is expressed in the cultivated tomato background, either by introgression of the cwp allele from primitive wild tomato species, such as Solanum habrochaites (cwph), or by transgenic overexpression, microfissures develop in the fruit cuticle, leading to the subsequent dehydration of the fruit. The importance of cwp is suggested by its ubiquitous expression in other members of the Solanaceae, e.g., tobacco (Nicotiana tabacum), pepper (Capsicum annuum), potato (Solanum tuberosum), and the green-fruited tomato wild relatives of tomato;3 however, the function of cwp is, yet, undetermined.
Over the years, horticultural growers of the dehydrating tomato have taken note that the severity of microfissures in fruit of cwph introgression lines was more pronounced when plants were grown in the greenhouse during the fall–winter (generally minimum night temperature of 15 °C) than when grown in spring–summer with higher day and night temperatures. This led us to study the role of low temperature on cwph-induced microfissuring, hypothesizing that there is a molecular–genetic component in the temperature effect on cwph.
It is well established that temperature changes affect gene expression in plants in general, and in tomatoes as well, leading to differential up- and downregulation of temperature-responsive genes22,23,24,25. In addition to the regulation of gene transcription, the role of temperature on alternative splicing of gene transcripts is also well established25,26,27,28, and the involvement of alternative splicing in response to abiotic stress has been the topic of exhaustive reviews29,30,31,32. Alternative splicing generates multiple mRNA and protein isoforms from a single gene and is common in all eukaryotes33,34. It was initially reported in plants nearly 30 years ago35 following earlier reports in animals. In plants, a significant number of intron-containing genes undergo alternative splicing process. More than half the intron-containing genes of Arabidopsis are subject to alternative splicing36,37,38, and a survey of nine species39 indicated that between 40 and 70% of the expressed intron-containing genes underwent alternative splicing. In an early study of tomato, Aoki et al.40 used cDNA library shotgun sequencing and found that only about 10% of the genes undergo alternative splicing, but more recently, Sun and Xiao27 by using HTS methods, showed that ca. 60% of the genes expressed in young tomato fruit exhibited alternative splicing. Most of alternative splicing events in plants are intron-retention type (61–68%), followed by alternative 3′ and alternative 5′ (25–28% together) and the rarest event of exon skipping (2–5%)41.
In this report, we show that the effect of low temperature on cuticular microfissuring and dehydration in cwp-expressing tomato fruit is via both the upregulation of cwp, compounded by alternative splicing of the cwp gene.
Materials and methods
Plant material
The segregating introgression line (SIL) for cwph and cwpe used in this study is as described in Hovav et al.3. The origin of the population is a backcross of the wild tomato species S. habrochaites LA1777 to the cultivated tomato (S. lycopersicum). A second segregating line, of miniature-sized plants for study in growth chambers, was developed from a BC3F2 of the cwph SIL to S. lycopersicum var. Microtom42 accompanied by genotyping for the cwph and cwpe alleles based on polymorphisms by using primers CWP-F 5′-CGTACTCAAACGATGATAAAGGT-3′ and CWP-R 5′-TTATTGCATTTGGAGTTTTTCAATCCG-3′.
Temperature treatments for whole plants
Plants were grown under standard conditions, as described previously43. For the growth of the Microtom-type (MT) plants under controlled temperature conditions, six plants of each of the cwp genotypes were grown in 500-ml pots until flowering. The flowering plants were transferred into two controlled growth chambers (Percival) set to temperatures 20/30 °C (night/day, high temperature) and 10/15 °C (night/day, low temperature). Flowers were marked at anthesis at the beginning of the temperature treatments. Following 21 days of incubation, five IG fruits from each plant were used for RNA extraction for cwp expression-level measurement, and the plants were transferred to a greenhouse for further development. Ripe fruits were picked from all plants, and the microfissure level was phenotyped as described below.
Temperature treatments for detached fruit
Temperature treatments were applied to detached IG fruit of either the SIL or the MT plants by placing the fruits under humid conditions in temperature-controlled chambers at temperatures (4, 10, 20, and 30 °C) and length of period (10 or 72 h), as described in this paper.
CWP expression level by quantitative PCR
The expression level of cwp was measured by quantitative PCR (qPCR) by using Maxima® SYBR-Green (Fermentas). In brief, first-strand cDNA was synthesized on template RNA that was extracted from tomato fruit pericarp. A qPCR reaction was conducted on 1 μl of cDNA as a template, with the primers qCWP-F 5′-CTATTTTCGTGGAAGTTGACAC-3′ and qCWP-R 5′-TGACAATTTGCTCTTTCCAC-3′ for amplification of CWP. The primers q18S-F 5′-GTGCATGGCCGTTCTTAGTTG-3′ and q18S-R 5′-CAGGCTGAGGTCTCGTTCGT-3′ were used for amplification of 18S rRNA as control. The relative expression level of cwp was calculated as the ratio of absolute expression of cwp to the absolute expression of 18S rRNA, by the equation
where CWPR is the relative expression level of CWP, CWPabs is the absolute expression level of CWP, and 18Sabs is the absolute expression level of 18S rRNA.
RNAseq of near-isogenic SIL fruit
For the study of global gene expression during fruit development, the SIL line segregants were grown, and fruit pericarp and peel were sampled from immature green (IG), mature green (MG), and breaker (Br) stages. For the study of the effect of temperature on detached fruitlets, IG fruitlets of MT BC3F7 lines were sampled and kept for 10 h, either at 20 or at 4 °C. Two replications, each of a minimum of three fruits, were prepared for each MT genotype. Tissues containing the skin and outer pericarp were sampled and immediately frozen in liquid nitrogen. Samples were prepared for RNAseq and run on Illumina HiSeq2000, as described in Shammai et al.44.
Bioinformatics—statistics and programs
Statistics of RNAseq
Raw reads were subjected to a filtering and cleaning procedure. The SortMeRNA tool45 was used to filter out rRNA, and the FASTX Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/index.html, version 0.0.13.2) was used to trim read-end nucleotides with quality scores <30, by using the FASTQ Quality Trimmer, and to remove reads with <70% base pairs with a quality score ≤30 by using the FASTQ Quality Filter. Statistics of the reads and quality are presented in Supplementary Table 1.
Mapping and expression analysis
Clean reads were aligned to the tomato genome extracted from the Sol Genomic Network database (https://solgenomics.net/organism/Solanum_lycopersicum/genome; SL3.0) by using Tophat2 software (v2.146). Gene abundance estimation was performed by using Cufflinks (v2.247) combined with gene annotations (ITAG3.10). Differential expression analysis was completed by using the edgeR R package48. Genes that varied more than twofold, with an adjusted p value of no more than 0.05, were considered differentially expressed. Venn diagrams were generated by using the online tool at bioinformatics.psb.ugent.be/webtools/Venn/. Functional annotation of the significantly expressed genes was prepared by using Plant MetGenMAP tool (http://bioinfo.bti.cornell.edu/cgi-bin/MetGenMAP/home.cgi). Heatmap visualization was performed by using R Bioconductor49.
Alternative splicing analysis
Intron abundance estimation was performed by using htseq-count (https://htseq.readthedocs.io/en/release_0.11.1/count.html) combined with gene annotations (ITAG3.10). Differential intron expression analysis was completed by using the edgeR R package.
Splice-related gene expression
Tomato splice-related genes were identified by using homology search (BLAST) against the Arabidopsis Splicing Related Gene Database (SRGD, http://www.plantgdb.org/SRGD/WB04.php, Wang and Brendel50).
Cloning the alternative transcripts of cwp
Total RNA from young tomato fruits of genotype cwph was extracted by using the Tri-Reagent (Invitrogene) according to the manufacturer’s instructions. First-strand cDNA was synthesized from the RNA template by using AMV reverse-transcriptase enzyme (Fermentas). The coding sequence (CDS) of the alternative transcripts was amplified by PCR reaction conducted on the cDNA, by using the primers CWP5UTR-F 5′-TCTTCATCTTATTCTTGTTTTTATTTATAG-3′ and the primer CWP3END-R 5′-TTATTGCATTTGGAGTTTTTCAATCCG-3′. The amplified fragments, comprising the seven alternative cwp transcripts, were cloned into pGEM®-Teasy plasmid (Promega) according to the manufacturer’s protocol. The plasmids were introduced into E. coli strain JM109 (Promega) by heat-shock treatment, and the bacteria planted on LB-agar plates containing 100 mg/l ampicillin and x-gal as selection markers. White colonies were subjected to PCR by using the universal primers SP6 and T7. Plasmids containing the sequences of the seven alternative variants were named pGEMAS1–pGEMAS7 (alternative transcripts VAR1– VAR7; correspondingly, sequences are presented in Supplementary File 1) and kept at −20 °C until further use.
Transgenic plants for the seven alternative transcripts
The coding sequence of the alternative transcripts was amplified by PCR reaction performed on the plasmids pGEMAS1– pGEMAS7, by using the primers CWPASNcoI-F that includes a restriction site for the restriction enzyme (RE) NcoI 5′-CATCCCATGGGTATAGTAGTGTTTATTTGGG-3′ (NcoI site is underlined), and CWPASSalI-R that includes a restriction site for the RE SalI 5′-CATGTCGACTTATTGCATTTGGAGTTTTTCAATCCG-3′ (SalI site is underlined). The PCR products were further cloned by using the pART7/pART27 binary system51. The resultant pART27 plasmids, under the control of 35S promoter, including the NptII gene (for kanamycin resistance) as marker selection, were introduced into Agrobacterium tumefaciens by using electroporation. Transgenic tomato plants (var. MP-1) were produced by the Agrobacterium method as described in ref. 3. Successful transformant plants were selected by PCR reaction conducted on the marker selection NptII, by using the primers NPTII-F 5′-TGAATGAACTGCAGGACGAG-3′ and NPTII-R 5′-AGCCAACGTATGTCCTGAT-3′. At least five independent T0 plants were selected for each of the alternative spliced coding sequences, and the selected plants were grown in a greenhouse and allowed to self-pollinate to produce T1 populations. T1 plants were grown as above, and mature green and ripe fruits were scored for microfissures and dehydration.
Quantitative measurement of alternative transcripts
Total RNA was extracted from detached IG tomato fruits of the SIL population, following incubation at three temperatures (10 , 20, and 30 °C) for 72 h. The cDNA was amplified by PCR by using the primers CWP5UTR-F and CWP3END-R as described above. PCR products were cloned to pGEM-Teasy vector, and the plasmids were introduced into E. coli strain JM109 by heat shock as described above. After transformation, bacteria were grown on agar plates containing 100 mg/l ampicillin and x-gal. At least 100 white colonies, each colony comprising a single transcription variant, were selected randomly for PCR reaction by using the universal primers SP6 and T7. The PCR products were separated on 2% agarose gel, together with the PCR products of the pGEMAS plasmids as size markers for the seven alternative transcripts. Number of each alternative transcript, measured by comparing its size to the size markers, was calculated based on the total alternative transcripts in three replications from each temperature treatment.
Results
Our previous results indicated that cwp is expressed in immature fruit of the green-fruited primitive wild species of tomato, including S. habrochaites, and in introgression lines derived from it harboring the habrochaites Solyc04g082540 ortholog, cwph. In contrast, the S. lycopersicon cwpe allele is not expressed. In this study, RNAseq results of developing tomato fruit of the segregating SIL corroborated this, showing that the cwph allele is expressed only transiently in the IG stage in fruit pericarp, while the cwpe allele is not expressed during development (Table 1).
The effect of temperature on cuticular microfissures
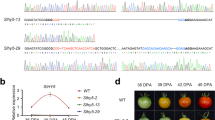
We observed over a number of seasons that the microfissure phenotype in cultivated tomato fruits of genotype cwph is generally more severe in winter-grown crops than in summer-grown crops. In the warmer weather, the fruits exhibit delicate and more sporadic microfissures, in comparison with winter. In order to study the effect of temperature during the early fruit development stage on microfissures, we developed miniature tomato plants of cwph and cwpe by backcrossing the cultivated tomato cwph-SIL to var. Microtom for three generations, selfing the heterozygous cwphe, and then selecting for the cwp allele genotypes by PCR. Six flowering plants of each genotype were grown in growth chambers for 3 weeks at low temperature (10/15 °C night/day) or high temperature (20/30 °C night/day) and then returned to a common greenhouse for continued development. Although both temperature conditions led to the microfissured cuticle on all fruits, significant differences were observed in the intensity of the microfissures. In response to low temperature, the fruits exhibited dense microfissures with deep scars, accompanied by suberization, while under the warm-temperature treatment, the fruits exhibited reduced microfissures and no scars were observed (Fig. 1).
Plants of genotype cwph in the MT background were grown in two ambient temperature ranges: low (10/15 °C night/day) and high (20/30 °C night/day) and tested for microcracking phenotype on the fruit cuticle. a Flowers were marked prior to the 2-week incubation (top), and the fruits continued to develop after the differential temperature incubation (bottom) in the greenhouse. b Representative ripe fruit from the temperature treatments (top, low temperature; bottom, high temperature). Incubation in low temperatures led to severer phenotype and deep cuticular “scars” (b, right top, arrow points to suberized scar) compared with high temperatures (b, right bottom)
Expression of the cwp h allele is upregulated under low temperature
We measured the expression of cwp in the IG fruit at the end of the 3-week temperature treatment and found that it was strongly upregulated by low temperature by about tenfold (Fig. 2a). In order to test whether the effect of low temperature on cwp expression is sensed by the fruit itself, rather than by the whole plant, we detached IG fruit of the two genotypes and maintained them at the same temperatures as above, for 72 h. The results showed that cwph gene expression in the detached fruit was increased about 15-fold in response to low temperature, similar to the whole plant (Fig. 2b). The silence of cwpe expression was not affected by the low temperature (not shown).
The left portion of the graph represents the expression level in IG fruitlets of whole plants following 21 days of temperature treatment. The right side of the graph indicates the expression of detached IG fruitlets treated for 72 h. Expression values are presented as relative to the high-temperature treatment in each experiment. Expression of cwpe fruitlets was nil
Global gene expression of developing fruit is affected by temperature
In order to further study the effect of low temperature on IG fruit gene expression, we performed a RNAseq study comparing global gene expression in detached IG fruits of the two cwp genotypes in the MT background, kept for 10 h at either 20 or 4 °C. The results further corroborated the upregulation of cwp gene expression in response to low temperature and further extended the temperature effect to the short time period of overnight (Table 2). Again, only the cwph allele is upregulated in response to low temperature; the cwpe allele remained silent and was not affected by low temperature.
The cwph gene is not unique with regard to being strongly regulated in response to low temperature. Whole-transcriptome analysis indicated significant differential regulation of transcription, and both genotypes showed temperature-dependent regulation. Fruit of the cwpe genotype had 2350 genes upregulated and 2642 genes downregulated by the low temperature, while the cwph genotype exhibited 3741 differentially regulated genes (1955 upregulated and 1786 downregulated), by at least twofold by low temperature. Common to both genotypes were 1290 upregulated, and 1249 downregulated, genes (Fig. 3, Supplementary Excel File 1). Between the two genotypes, a total of ca. 6000 genes were differentially expressed in response to temperature, comprising ca. 25% of the expressed genome.
cwp h undergoes alternative splicing and produces seven alternative variants
PCR of the cwph cDNA indicated multiple alternative transcripts (Supplementary Fig. 1a). Sequencing of the multiple PCR products indicated seven alternative transcripts of the cwp gene, termed VAR1–VAR7 (Fig. 4). VAR1 was the longest sequence, the product of the removal of three introns and retention of four exons. In comparison with VAR1, the other variants are a result of four different alternative splicing events: intron 2 retention, alternative 3′ acceptor of intron 2, alternative 5′ donor of intron 1, and exon 2 skipping. VAR2 undergoes an intron 2 retention, VAR3 is processed via alternative 3′ acceptor of intron 2, VAR4 results from a combination of alternative 3′ acceptor of intron 2 and alternative 5′ donor of intron 1, VAR5 undergoes alternative 5′ donor of intron 1, VAR6 is formed by a combination of alternative 3′ acceptor of intron 2, alternative 5′ donor of intron 1, and exon 2 skipping, and VAR7 is formed by a combination of intron 2 retention and alternative 5′ donor of intron 1. All the introns of the cwph alternative splicing variants are U2 type, and follow all the U2-type intron rules: (1) the sequence AG/GUAAGU (exon/intron, underlined are the conserved bases GU) at the 5′ junction of the intron, (2) the sequence UGYAG/GU (intron/exon, underlined are the conserved bases AG) at the 3′ junction of the intron, (3) U/A-rich regions, (4) a conserved YURAY sequence in the branchpoint, 30–70 bp upstream to the 3′ junction of the intron, and (5) a poly-U tract between the YURAY sequence and the 3′ junction of the intron (Supplementary Fig. 2).
Interestingly, when cwph cDNA from wild species S. habrochaites fruit, as well as from other primitive species that express the cwp gene, was analyzed for alternative transcripts, only three were observed, compared with the seven produced in the S. lycopersicon background of the introgression line (Supplementary Fig. 1b). This suggests that the pre-mRNA of cwph may be differentially spliced, depending on the endogenous splicing machinery, which likely differs between the wild species and the cultivated species.
The seven alternative transcripts comprise reading frames for five proteins
The reading frames of the alternative transcripts of cwph differ significantly in length (Fig. 4, Supplementary Fig. 2, Supplementary Table 2). The longest reading frame was found in VAR1 with 811 bp in length coding for a 270 aa protein. VAR2 is the longest variant with 889 bp but a reading frame for a shortened protein of 187 aa, due to a premature stop codon in exon 3. The length of VAR3 is 666 bp with a reading frame for a 185-bp protein, also due to a premature stop codon in exon 3. VAR4, VAR5, and VAR7 share the same reading frame of 73 aa due to a premature stop codon in exon 2. VAR6 is dramatically shorter than the other variants due to its unique exon 2 skipping, but its reading frame encodes for a 122 aa protein. In total, five different potential proteins were identified, products of the seven alternative transcripts. All five proteins share a 73 aa N terminus, while the three longest proteins (products of VAR1, VAR2, and VAR3) share a longer region of 158 aa at the N terminus. VAR6 contains the 73 aa at the N terminus, and shares the C terminus with VAR1, due to the reversion to the reading frame (Fig. 4b).
Functional analysis of alternative VARs
In order to determine which of the different splice variants are functional, we developed transgenic tomato plants for each (except VAR5 that shares the same reading frame as VAR4 and VAR7) in the background of variety MP-1, which we previously used to functionally express the cwp gene3. The results clearly showed that only transgenic VAR1 and VAR2 exhibited microfissures and subsequent dehydration. The other variants, as well as the control MP-1 fruit, did not show any cwph-related phenotypes, neither microfissures nor dehydration (Fig. 5a).
a Microfissures were observed on the fruit peel of variants VAR1 and VAR2 only (upper row), and VAR1 and VAR2 were the only variants exhibiting dehydration after 15 days. b VAR2 itself undergoes alternative splicing, and PCR on VAR2 transgenic tomato results in three bands, VAR1, VAR2, and VAR3. c A schematic illustration of the alternative splicing events in intron 2 that leads to the production of VAR1 and VAR3 from VAR2
Further analysis of the functional transgenic VAR2 showed that VAR2 itself undergoes alternative splicing, yielding the VAR1 gene product (Fig. 6). Since VAR2 is produced as a result of intron 2 retention, intron 2 can still be removed from VAR2, either to produce VAR1 or by the alternative 3′ process to produce VAR3. PCR amplification of the cDNA of transgenic VAR2 indeed resulted in three bands, corresponding to the three alternative transcripts VAR1, VAR2, and VAR3 (Fig. 5b, c). Thus, we conclude that the longest ORF leading to the 270-amino acid CWP protein is the functional protein.
a Products of PCR reaction performed on cwp cDNA after incubation in 10 (lanes 1–3), 20 (lanes 4–6), and 30 °C (lanes 7–9). Lane 10 is a non-template control, lane 11 is a size marker. The arrows point to PCR products significantly affected by temperature. b The relative ratio of the alternative transcripts after incubation in the three temperatures
Low temperature changes the pattern of alternative splicing leading to a further increase in the functional VAR1
In order to determine whether low temperature also impacts on the pattern of alternative splicing of cwph, we quantified the different transcript variants in response to temperature treatment of detached IG fruitlets (10, 20, or 30 °C). The alternative transcripts of cwph were quantified following PCR cloning of the fruit cDNA and determining transcript size from each treatment in 100 transformed E. coli colonies. The results indicated a significant shift in the ratio between two alternative transcripts; low temperature increased the relative proportion of the functional VAR1 transcripts, at the expense of the nonfunctional VAR3 (Fig. 6). Under high temperature, VAR1 accounted for 38% of the transcripts, and this increased to more than 50% under low temperature. VAR3 made up 29% of the transcripts in higher temperature but only 10% in the lower temperature. The remaining variants were present at lower levels and were largely unaffected by temperature. An interesting observation is the increase in VAR5 under low temperature, analogous to the response of VAR1. VAR1 and VAR5 share the alternative splice site, suggesting that this splice site may be specifically affected by temperature.
Global gene alternative splicing is modified by low temperature
Just as cwp is not the only gene differentially expressed due to temperature, so too it is not unique in showing alternative splicing in response to temperature. Following the mapping of the RNAseq reads and subsequent FPKM analysis based on the annotated coding sequences of each gene, the reads were similarly mapped specifically to the intron regions of the annotated tomato genome. Thus, we could determine whether temperature affected the expression of intronic regions, serving as an indication of alternative splicing. For the cwph genotype, we observed that 4091 genes showed differential expression of intronic regions (>2-fold, padj <0.05) between the two temperature treatments. These data include any significant change in the number of intron-mapped reads per gene. Of the 3741 genes differentially expressed in the cwph genotype in response to temperature (Fig. 3), 1601 were also characterized by differential expression of intron regions (Fig. 7). An equal number, about 800, of both low-temperature upregulated and downregulated genes showed differential intron retention.
Splicing-related genes are differentially expressed under the influence of temperature
In order to shed light on the cause of the temperature-dependent differential alternative splicing, we analyzed the expression data for the known splicing-related genes in the tomato genome. The tomato homologs of the Arabidopsis splicing-related genes were identified by BLAST against the Arabidopsis database (SRGD, http://www.plantgdb.org/SRGD/WB04.php, Wang and Brendel50), and 174 tomato genes that showed at least 60% identity to one of the Arabidopsis genes were selected (Supplementary Excel file 2). Analysis of the expression data for these 174 genes indicated that 29 splicing-related genes were differentially expressed (>2-fold, padj <0.05) in response to the low temperature in both allelic genotypes (Fig. 8, Supplementary Excel file 3). Ten genes were upregulated by the low temperature and 19 were downregulated irrespective of the cwp genotype.
Heat map of the 29 splicing-related genes differentially expressed (>2-fold, padj <0.05) in response to temperature. List of the tomato homologs of the Arabidopsis splicing-related genes are listed in Supplementary Excel file 2. Fruitlets of the two genotypes (cwph, HH; cwpe, EE) were subjected to either 4 or 20 °C for 10 h
Discussion
The wild species S. habrochaites allele cwph introgressed into the cultivated S. lycopersicon leads to the development of microfissures in the tomato fruit cuticle, leading to its subsequent dehydration3. In contrast, the cultivated tomato is relatively impervious to water loss, due to a continuous fruit cuticle. Accordingly, the evolutionary silencing of cwp expression during Solanum evolution is one of the factors that contributed to the evolution under domestication of the fleshy tomato fruit. Our study accounts for the increased severity of microfissuring observed under low temperatures. Both transcriptional and post-transcriptional components of cwp gene expression were found to be affected by low temperature.
The relationship between the expression of functional splice variants of the cwp gene and the phenotype of microfissures was tested under a range of temperature conditions. A strong correlation was demonstrated between the temperature-modulated cwp expression and the intensity of cuticular microfissure, with low temperature causing upregulation of cwp expression and severe microfissures, while high temperature leads to reduced cwp expression and mild microfissures. In addition, there is a further increase in the relative proportion of the VAR1 splice variant, under low temperatures, and we show that it is the functional variant, thereby compounding the effect of low temperature on the severity of the microfissures.
While the expression of the cwph allele is strongly upregulated by low temperature, the cwpe allele remains silent. Interestingly, the cwp gene is the only such gene that is silent in the cwpe genotype, irrespective of temperature, and upregulated significantly in the cwph genotype in response to low temperature.
Ambient temperature is a well-established factor controlling gene expression in plants. Our results indicate that in developing tomato fruit, low temperature of 4 °C for 10 h is perceived by the fruit tissue per se and leads to the differential expression of ~ 6000 of the expressed genes, or ca. 25% of the tomato genome. This is in general agreement with other studies, including those of tomato tissue. For example, Chen et al.25 similarly reported about 6000 genes differentially expressed in tomato leaf tissue following 12 h at 4 °C.
Alternative splicing plays an important role in adaptation of plants to abiotic stress and environmental constraints22,52,53. Evidence for the effect of temperature stresses on the plant transcriptome was shown by Filichkin et al.36, who observed not only an altered abundance of many transcripts but also changes in their alternative splicing pattern. More recently, this area of study has benefitted from HTS in showing the temperature effects on alternative splicing patterns, including in tomato25,26,27,28. While the effect of low temperature on alternative splicing patterns is a global transcriptome phenomenon, the physiological impact is likely due to individual gene transcript modifications. One of the earliest reports showing the effect of low temperature on a physiological process via alternative splicing of a particular gene was the study of the low-temperature effect on a potato invertase gene related to cold-sweetening during potato storage54. In this study, temperature impacted on the retention of the smallest known plant exon, 9 bp, which translates to the NDP catalytic domain, with concomitant implications on sucrose hydrolysis.
Our results with respect to the occurrence of alternative splicing are also in general agreement with other studies. Particularly regarding young tomato fruit, Sun and Xiao27 reported that AS characterizes 60% of multi-exon genes during young tomato fruit development, similar to our results. Since our RNAseq reads were single-end 50 bp (SE50), the ability to fully characterize alternative splicing was hampered. Therefore, our observation was based on the differential expression of introns and is not a strict analysis of alternative splicing. Nevertheless, our results clearly indicate that cwp is not the only differentially expressed gene that also undergoes alternative splicing. Not all AS genes affected by temperature are also differentially expressed, and in 2490 genes temperature affects only the variant splicing mechanism. Reciprocally, 2140 genes are differentially expressed but do not show differential intron expression. cwp is one of about 1600 genes that are both differentially expressed and alternatively spliced in response to low temperature, However, in the case of cwp, this leads to a synergistic effect on the production of functional processed mRNA and hence the horticultural phenotype.
The effect of temperature on alternative splicing is an established phenomenon30,31,55, and the response to temperature is rapid56, likely occurring in even less than the 10-h treatment we report. Significantly, the splicing mechanism itself is affected by temperature. Verhage et al.57 recently reported that even under modest temperature changes, there were significant effects on splicing-related genes, suggesting that the whole spliceosome is sensitive to temperature. They present a two-step model of temperature control of plant development: first, the alternative splicing of splicing-mechanism genes, followed by the subsequent alternative splicing of downstream genes, including genes of the transcriptional network25,55. To this, we might add a further initial step to the model based on our results: the differential regulation of gene expression of the splicing-mechanism genes, impacting on the reservoir for alternative splicing variants.
The important contribution of the endogenous splicing machinery is emphasized particularly in the introgression lines. While the endogenous cwph allele in S. habrochaites fruit showed three alternative transcripts, the cwph introgressed in the S. lycopersicon background that expressed seven variants. This suggests a genetic background effect for the alternative splicing process, based perhaps on genetic variability between the species in splicing-mechanism genes, either due to genetic variability in their biochemical function or to differences in expression or its own alternative splicing.
We show that the functional variant with respect to microfissures is the VAR1, with the longest mRNA product. The other variants are characterized by premature stop codons that lead to shortened reading frames and truncated proteins. These RNAs may likely be targeted to rapid degradation through the nonsense-mediated-decay (NMD) pathway58,59. As such, the case of cwp is an example of alternative splicing exhibiting physiological significance32. Nevertheless, we cannot conclude with certainty that in fact the VAR1 is the “functional” variant since we do not yet know what the physiological function of cwp actually is. While cwp is expressed in the green-fruited wild species of tomato3, these wild species fruit do not exhibit microfissures7, indicating that the physiological role of cwp is not in developing microfissures per se. Rather, it is only in the background of the cultivated tomato that the phenotype of microfissures is observed when cwp is expressed. One could hypothesize that cuticular microfissuring is an unrelated phenotype produced serendipitously by the interaction of cwp with S. lycopersicon components of fruit rind development, and that the physiological role of cwp in the primitive wild species and other Solanaceae is to be found elsewhere. Thus, it is still possible that the other alternative transcripts do indeed have a function, perhaps even the natural physiological one60. The unknown function of cwp remains to be revealed, and the effect of low temperature on upregulating its expression may be useful in this pursuit.
References
Jeffree, C. E. in Biology of the Plant Cuticle (eds Riederer, M. and Muller, C.) 11–125 (Blackwell Publishing Ltd, Oxford, UK, 2006).
Nawrath, C. Unraveling the complex network of cuticular structure and function. Curr. Opin. Plant Biol. 9, 281–287 (2006).
Hovav, R., Chehanovsky, N., Moy, M., Jetter, R. & Schaffer, A. A. The identification of a gene (Cwp1), silenced during solanum evolution, which causes cuticle microfissuring and dehydration when expressed in tomato fruit. Plant J. 52, 627–639 (2007).
Bargel, H. & Neinhuis, C. Tomato (Lycopersicon esculentum Mill.) fruit growth and ripening as related to the biomechanical properties of fruit skin and isolated cuticle. J. Exp. Bot. 56, 1049–1060 (2005).
Wilson, L. A. & Sterlin, C. Studies on the cuticle of tomato fruit, I. fine structure of the cuticle. Zeit Pflanz. 77, 359–371 (1976).
Spooner, D. M., Peralta, I. E. & Knapp, S. Comparison of AFLPs with other markers for phylogenetic inference in wild tomatoes [Solanum L. section Lycopersicon (Mill.) Wettst.]. Taxon 54, 43–61 (2005).
Yeats, T. H. et al. The fruit cuticles of wild tomato species exhibit architectural and chemical diversity, providing a new model for studying the evolution of cuticle function. Plant J. 69, 655–666 (2011).
Peet, M. M. Fruit cracking in tomato. HortTechnology 2, 216–223 (1992).
Dorais, M. D. A., Dermers, W., van Iepern, S. & Papadoupoulos, A. P. Cuticle cracking in tomato fruit. Hort. Rev. 30, 163–184 (2004).
Hen-Avivi, S., Lashbrooke, J., Costa, F. & Aharoni, A. Scratching the surface, genetic regulation of cuticle assembly in fleshy fruit. J. Exp. Bot. 65, 4653–4664 (2014).
Isaacson, T. et al. Cutin deficiency in the tomato fruit cuticle consistently affects resistance to microbial infection and biomechanical properties, but not transpirational water loss. Plant J. 60, 363–377 (2009).
Jiang, F. et al. Disassembly of the fruit cell wall by the ripening-associated polygalacturonase and expansin influences tomato cracking. Hortic. Res. 6, 17 (2019).
Lashbrooke, J. et al. The tomato MIXTA-like transcription factor coordinates fruit epidermis conical cell development and cuticular lipid biosynthesis and assembly. Plant Physiol. 169, 2553–2571 (2015).
Leide, J., Hildebrandt, U., Reussing, K., Riederer, M. & Vogg, G. The developmental pattern of tomato fruit wax accumulation and its impact on cuticular transpiration barrier properties, effects of a deficiency in a beta-ketoacyl-coenzyme A synthase (LeCER6). Plant Physiol. 144, 1667–1679 (2007).
Moctezuma, E., Smith, D. L. & Gross, K. C. Antisense suppression of a beta-galactosidase gene (TB G6) in tomato increases fruit cracking. J. Exp. Bot. 54, 2025–2033 (2003).
Adams, S. R., Cockshull, K. E. & Cave, C. R. J. Effect of temperature on the growth and development of tomato fruits. Ann. Bot. 88, 869–877 (2001).
Baker, E. A., Bukovac, M. J. & Hunt, G. M. in The Plant Cuticle (eds Culter D. F., Alvin K. L., Price, C. E.) 33–44 (Academic Press, London, 1982).
Domınguez, E., Cuartero, J. & Heredia, A. An overview on plant cuticle biomechanics. Plant Sci. 18, 77–84 (2011).
Domínguez, E. et al. Tomato fruit continues growing while ripening, affecting cuticle properties and cracking. Physiol. Plant. 146, 473–486 (2012).
España, L. et al. Biomechanical properties of the tomato (Solanum lycopersicum) fruit cuticle during development are modulated by changes in the relative amounts of its components. New Phytol. 202, 790–802 (2014).
Segado, P., Domínguez, E. & Heredia, A. Ultrastructure of the epidermal cell wall and cuticle of tomato fruit (Solanum lycopersicum l.) during development. Plant Physiol. 170, 935–946 (2016).
Knight, M. R. & Knight, H. Low-temperature perception leading to gene expression and cold tolerance in higher plants. New Phytol. 195, 737–751 (2012).
Lurie, S., Handos, A., Fallik, E. & Shapira, R. Reversible inhibition of tomato fruit gene expression at high temperature effects on tomato fruit ripening. Plant Physiol. 110, 1207–1214 (1996).
Sanchez-Bel, P. et al. Proteome changes in tomato fruits prior to visible symptoms of chilling injury are linked to defensive mechanisms, uncoupling of photosynthetic processes and protein degradation machinery. Plant Cell Physiol. 53, 470–484 (2012).
Chen, H. et al. A comparison of the low temperature transcriptomes of two tomato genotypes that differ in freezing tolerance, Solanum lycopersicum and Solanum habrochaites. BMC Plant Biol. 15, 132 (2015).
Keller, M. et al. Alternative splicing in tomato pollen in response to heat stress. DNA Res. 24, 205–217 (2017).
Sun, Y. & Xiao, H. Identification of alternative splicing events by RNA sequencing in early growth tomato fruits. BMC Genomics 16, 948 (2015).
Wang, G., Weng, L., Li, M. & Xiao, H. Response of gene expression and alternative splicing to distinct growth environments in tomato. Int. J. Mol. Sci. 18, E475 (2017).
Laloum, T., Martin, G. & Duque, P. Alternative splicing control of abiotic stress responses. Trends Plant Sci. 23, 140–150 (2018).
Mastrangelo, A. M., Marone, D., Laidò, G., De Leonardis, A. M. & De Vita, P. Alternative splicing, enhancing ability to cope with stress via transcriptome plasticity. Plant Sci. 185-186, 40–49 (2012).
Shang, X., Cao, Y. & Ma, L. Alternative splicing in plant genes, a means of regulating the environmental fitness of plants. Int. J. Mol. Sci. 18, E432 (2017).
Carvalho, R. F., Feijão, C. V. & Duque, P. On the physiological significance of alternative splicing events in higher plants. Protoplasma 250, 639–650 (2013).
Lareau, L. F., Green, R. E., Bhatnagar, R. S. & Brenner, S. E. The evolving roles of alternative splicing. Curr. Opin. Struct. Biol. 14, 273–282 (2004).
Stamm, S. et al. Function of alternative splicing. Gene 344, 1–20 (2005).
Werneke, J. M., Chatfield, J. M. & Ogren, W. L. Alternative mRNA splicing generates the two ribulosebisphosphate carboxylase/oxygenase activase polypeptides in spinach and Arabidopsis. Plant Cell 1, 815–825 (1989).
Filichkin, S. A. et al. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 20, 45–58 (2010).
Marquez, Y., Brown, J. W. S., Simpson, C., Barta, A. & Kalyna, M. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 22, 1184–1195 (2012).
Reddy, A. S., Marquez, Y., Kalyna, M. & Barta, A. Complexity of the alternative splicing landscape in plants. Plant Cell 25, 3657–3683 (2013).
Chamala, S., Feng, G., Chavarro, C. & Barbazuk, W. B. Genome-wide identification of evolutionarily conserved alternative splicing events in flowering plants. Front. Bioeng. Biotechnol. 3, 33 (2015).
Aoki, K. et al. Large-scale analysis of full-length cDNAs from the tomato (Solanum lycopersicum) cultivar Micro-Tom, a reference system for the Solanaceae genomics. BMC Genomics 11, 210 (2010).
Ner-Gaon, H., Leviatan, N., Rubin, E. & Fluhr, R. Comparative cross-species alternative splicing in plants. Plant Physiol. 144, 1632–1641 (2007).
Meissner, R. et al. A new model system for tomato genetics. Plant J. 12, 1465–1472 (1997).
Miron, D. & Schaffer, A. A. Sucrose phosphate synthase, sucrose synthase, and invertase activities in developing fruit of Lycopersicon esculentum Mill. and the sucrose accumulating Lycopersicon hirsutum Humb. and Bonpl. Plant Physiol. 95, 623–627 (1991).
Shammai, A. et al. Natural genetic variation for expression of a SWEET transporter among wild species of Solanum lycopersicum (tomato) determines the hexose composition of ripening tomato fruit. Plant J. 96, 343–357 (2018).
Kopylova, E., Noé, L. & Touzet, H. SortMeRNA, fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28, 3211–3217 (2012).
Kim, D. et al. TopHat2, accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R3–R6 (2013).
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR, a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Gentleman, R. C. et al. Bioconductor, open software development for computational biology and bioinformatics. Genome Biol. 5, R80 (2004).
Wang, B. B. & Brendel, V. Genome wide comparative analysis of alternative splicing in plants. Proc. Natl Acad. Sci. USA 103, 7175–7180 (2006).
Gleave, A. P. A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20, 1203–1207 (1992).
Dubrovina, A. S., Kiselev, K. V. & Zhuravlev, Y. N. The role of canonical and noncanonical pre-mRNA splicing in plant stress responses. Biomed. Res. Int. 2013, 264314 (2013). Article ID.
Staiger, D. & Brown, J. W. Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 25, 3640–3656 (2013).
Bournay, A. S., Hedley, P. E., Maddison, A., Waugh, R. & Machray, G. C. Exon skipping induced by cold stress in a potato invertase gene transcript. Nucleic Acids Res. 24, 2347–2351 (1996).
Seo, P. J., Park, M. J. & Park, C. M. Alternative splicing of transcription factors in plant responses to low temperature stress, mechanisms and functions. Planta 237, 1415–1424 (2013).
Calixto, C. P. G. et al. Rapid and dynamic alternative splicing impacts the arabidopsis cold response transcriptome. Plant Cell 30, 1424–1444 (2018).
Verhage, L. et al. Splicing-related genes are alternatively spliced upon changes in ambient temperatures in plants. PLoS ONE 12, e0172950 (2017).
Kalyna, M. et al. Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 40, 2454–2469 (2011).
Ohtani, M. & Wachter, A. NMD-based gene regulation - a strategy for fitness enhancement in plants? Plant Cell Physiol. 60, 1953–1960 (2019).
Filichkin, S., Priest, H. D., Megraw, M. & Mockler, T. C. Alternative splicing in plants, directing traffic at the crossroads of adaptation and environmental stress. Curr. Opin. Plant Biol. 24, 125–135 (2015).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chechanovsky, N., Hovav, R., Frenkel, R. et al. Low temperature upregulates cwp expression and modifies alternative splicing patterns, increasing the severity of cwp-induced tomato fruit cuticular microfissures. Hortic Res 6, 122 (2019). https://doi.org/10.1038/s41438-019-0204-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41438-019-0204-9
This article is cited by
-
The SlSHN2 transcription factor contributes to cuticle formation and epidermal patterning in tomato fruit
Molecular Horticulture (2022)
-
Alternative splicing during fruit development among fleshy fruits
BMC Genomics (2021)