Abstract
Purpose
Secondary findings (SFs) are present in 1–4% of individuals undergoing genome/exome sequencing. A review of how SFs are disclosed and what outcomes result from their receipt is urgent and timely.
Methods
We conducted a systematic literature review of SF disclosure practices and outcomes after receipt including cascade testing, family and provider communication, and health-care actions. Of the 1,184 nonduplicate records screened we summarize findings from 27 included research articles describing SF disclosure practices, outcomes after receipt, or both.
Results
The included articles reported 709 unique SF index recipients/families. Referrals and/or recommendations were provided 647 SF recipients and outcome data were available for 236. At least one recommended evaluation was reported for 146 SF recipients; 16 reports of treatment or prophylactic surgery were identified. We found substantial variations in how the constructs of interest were defined and described.
Conclusion
Variation in how SF disclosure and outcomes were described limited our ability to compare findings. We conclude the literature provided limited insight into how the American College of Medical Genetics and Genomics (ACMG) guidelines have been translated into precision health outcomes for SF recipients. Robust studies of SF recipients are needed and should be prioritized for future research.
Similar content being viewed by others
INTRODUCTION
Due to their increasing diagnostic utility and affordability, genome and exome sequencing have become widely used. An estimated 1–4% of individuals undergoing genome or exome sequencing have a medically actionable variant unrelated to the indication for sequencing [1,2,3,4]. The American College of Medical Genetics and Genomics (ACMG) first recommended “opportunistic screening” for medically actionable secondary findings (SFs) in 2013 [5]. Opportunistic screening is defined as analysis of acquired data unrelated to the indication of the test for the purpose of detecting unrecognized disease risk. A 2017 update of these guidelines recommends pathogenic and likely pathogenic variants in 59 genes associated with 27 genetic disorders be returned [6]; an additional update was recently published [7]. In addition to the clinical testing setting, some research initiatives (including some preceding release of the first set of ACMG guidelines), addressing ethical imperatives [8] to return medically actionable variants to their participants, base decisions on which variants to return on the ACMG guidelines [2, 9,10,11]. ACMG™, ACMG SF™, ACMG 59™, ACMG 56™, and related words and designs incorporating ACMG™, are trademarks of the American College of Medical Genetics and Genomics and may not be used without permission.
Understanding both the implementation and the impact of the ACMG SFTM policy is an urgent priority. Numerous position statements, ethics reviews, and thought pieces have been written about opportunistic return of actionable genomic findings (including recent disagreement between the US and European contexts) [9, 11,12,13,14,15]. Despite this, fundamental questions such as the prevalence of disease in SF recipients and their family members and the impact of SF return on morbidity and mortality remain unanswered [3]. Understanding adherence to health behavior and family communication recommendations related to SF is a key to answer these questions. As large-scale precision health genomics studies become more routine, an increasing number of SFs meeting ACMG criteria will be disclosed in the coming years [16, 17]. Lessons learned from opportunistic screening for SFs will provide valuable insights to guide the development of population-level genomic screening efforts. We undertook this systematic literature review to understand the state of the science surrounding disclosure practices in opportunistic screening of medically actionable SFs and outcomes associated with their receipt.
MATERIALS AND METHODS
Our goal with this review was to evaluate the evidence base describing the following key elements of the implementation of the ACMG SFTM return policy: (1) SF disclosure practices, including personnel involved, recommendations communicated, and strategies employed, or (2) outcomes associated with SF receipt including psychological impact, communication, health-care behaviors, cascade testing of relatives, and health outcomes. To do this, we analyzed reports of any of these elements at the individual SF recipient level across all included literature.
Search strategy
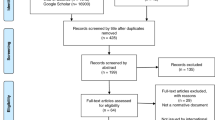
We conducted a systematic search for publications addressing at least one of our objectives in specific patients or research study participants per PRISMA procedures [18] (Fig. 1). Searches were conducted across six databases (PubMed, Web of Science, PsychINFO, CINAHL, Embase, and Scopus) and in the gray literature (academic papers, including theses and dissertations, research and committee reports, government reports, conference papers, and ongoing research, among others) from 1 January 2012 through 31 July 2020. An iterative process was used to select search terms for this review, including review of several exemplar articles and snowball techniques [19]. Search terms used to describe SF were combined with terms describing sequencing methodologies (e.g., exome sequencing, genome sequencing) and terms describing processes and outcomes (see Supplementary Materials and Methods for the complete search strategy employed with each database). Several gray literature sources, including Google Scholar, the New York Academy of Medicine’s grayLit.org database, and the Canadian Agency for Drugs and Technologies in Health were also searched. A broader and less specific search strategy was employed for gray literature sources; however, these sources either duplicated the findings of the database searches (e.g., Google Scholar) or did not yield any references that were included in the analysis and this process is thus not represented graphically.
The protocol for this review was not prospectively registered because the review coincided with significant updates to the PROSPERO registration process.
Article selection
Primary research articles reporting longitudinal and cross-sectional qualitative and quantitative data derived from case reports, case–control/case series, observational cohort studies, and other research modalities were included in this review. Although reporting ACMG SFTM is recommended for individuals undergoing clinical exome/genome sequencing, reports of variants meeting ACMG criteria returned to individuals from research sequencing, and clinical and research panel-based testing, were included provided the variant was germline and the other criteria in Table 1 were met to capture as much data as possible. Because SFs are uncommon, articles where SF disclosure practices and/or outcomes were not a primary or major focus were included if sufficient information was provided within the main body of the article or in the Supplemental material to accomplish our objectives. The criteria used to define SFs for purposes of this review are outlined in Table 1. While we did not perform individual pathogenicity assessments, descriptions of reported variants were reviewed to ensure consistency with our definition.
Articles were included if they described how a qualifying SF was disclosed to a specific research participant, healthy volunteer, patient, or parent of a pediatric proband, and/or if they described any actions taken by recipients of a SF and/or their relatives or any psychological reaction to receiving such a result. Articles were excluded if (1) they were not published in English, (2) only described methods to assess pathogenicity or laboratory reporting practices associated with SF, (3) only described the incidence/prevalence of SF in a particular cohort, (4) focused primarily on the ethics or policy implications of sequencing/return of results, (5) only assessed or described hypothetical results, or (6) or if disclosure or outcome data for qualifying SF were aggregated with additional genomic variants in such a way that disentangling outcomes and/or disclosure practices specific to SF was impossible.
Two authors (J.C.S. and F.M.F.) independently performed title and abstract screening of all nonduplicate records using the eligibility criteria described above. Records selected for full-text review were read by both authors and discrepancies regarding articles selected for final inclusion were resolved in a series of phone meetings. Excluded articles assigned were assigned to the exclusion categories shown in Fig. 1. Articles assigned (either during full-text review or title/abstract screening) to the Ethics/Commentary and Stakeholder Attitudes and Preferences Only exclusion categories by one but not both of the two primary reviewers were then reviewed by a third author (E.M.) who resolved discrepancies.
Data extraction and analyses
Bibliographic information, study characteristics, and findings were extracted independently by J.C.S. and F.M.F. using a data extraction form in Excel. An individual receiving a SF comprised the unit of analysis for this review. Disclosure practices and/or outcomes described at the SF recipient level were extracted along with the total number of SF recipients reported in each included article. A framework analysis approach was employed to thematically categorize and organize data across studies [20]. The two primary reviewers examined the complete set of included articles to create an analytical framework centered on each of the major foci of this review: SF disclosure practices and outcomes reported after SF receipt. Major themes from each topic were identified and data iteratively analyzed and integrated into the framework. F.M.F. performed the initial coding of results pertaining to the “disclosure practices” theme and J.C.S. performed the initial coding pertaining to the “outcomes” theme; each author manually reviewed the other’s work. Additional articles included as a result of the final updated search were analyzed using this framework. The final thematic data resulting from this process were synthesized to generate the conclusions.
Critical appraisal
Critical appraisal of the included articles was not performed because the topics of interest for this review were rarely the primary focus of the publication. PRISMA standards recommend assessing articles for bias in individual studies and across studies. Bias is difficult to assess when articles are describing data gathered as other than the primary design of a study. We recognize that undetected biases may have been present in the articles described here. In at least two instances, information pertaining to the topics of this review was not found in the main body of the paper but was detailed in the Supplemental material.
RESULTS
Inclusion characteristics
Twenty-seven published articles met criteria for inclusion in this review (Table 2). The primary focus of 13 of the included articles was to systematically and specifically describe SF disclosure practices and/or outcomes in SF recipients, whereas others described these processes and/or outcomes for a broader range of genetic variants including SFs (n = 9). Six of the articles described SFs from clinical sequencing [21,22,23,24,25,26] and three of these were clinical reports describing a single patient/family. The remainder described SFs obtained via sequencing conducted as part of a research study (n = 18) or biobank (n = 4). All but eight of the articles originated in the United States. Seven articles did not report any outcome data. Four articles described panel-based sequencing efforts [23, 24, 27, 28]. See Table 3 for additional characteristics of the included articles.
Records that did not meet inclusion criteria either at the title/abstract or full-text screening stages were grouped into exclusion categories. Of the 1,157 excluded records, 164 focused primarily on the attitudes, preferences, opinions, or perspectives of various stakeholders surrounding the return of genomic sequencing results and 283 were editorials, commentaries, or research studies primarily focused on the ethics and legal issues of returning genomic sequencing results.
Thematic analysis and summary data
Our analysis framework identified five major themes relating to SF disclosure practices and seven major themes related to outcomes associated with SF receipt (Table 2) in addition to cost estimates associated with SF return. A total of 709 SF index recipients/families were reported; outcomes data were only available for 33% (n = 236) of them.
Nine articles reported data on more than ten SF recipients [1, 4, 21, 29,30,31,32,33,34]. One did not report any disclosure practice data [21] and two were publications describing the same group of SF recipients, participants in a large US biobank study [29, 33]. This pair of studies provided detailed disclosure practice data for a cohort of 515 SF recipients [33]; the uptake of recommended health-care actions in the one-year period following disclosure among 48 of these individuals who had received BRCA1/2 SF were evaluated via records review [29]. Four articles described standardized SF disclosure to participants in research studies; two of these enrolled healthy individuals [30, 32], one enrolled patients with cancer [31], and the other enrolled patients affected with a variety of conditions [4]. One of these studies standardized the interval between disclosure and evaluation of postdisclosure outcomes (four months) [4]; this interval varied within the remaining studies with a range from three months to four years. One article [1] described 66 SF recipients from a research sequencing consortium comprising nine different studies; neither SF disclosure nor the related outcomes studied were standardized across sites [35]. Most of the data reported in this article was derived from retrospective case reports submitted by the study sites although 18 SF recipients completed a follow-up interview using an interview guide harmonized with another included study [4]. Finally, in one article describing outcomes in 11 SF recipients, disclosures were not standardized and the extent to which management recommendations were provided to SF recipients was not clear [34].
Disclosure practices
Personnel involved in disclosure
Seventeen articles described the specific personnel involved in the disclosure of a SF to a total of 618 SF recipients [4, 23, 25, 27, 30,31,32,33,34, 36,37,38,39,40,41,42,43]. Genetic counselors and medical geneticists were the disclosing providers in ten articles [4, 23, 31, 32, 34, 36, 39,40,41,42] and three additional articles described a team approach to disclosure that included personnel with genetics expertise [25, 27, 38]. A cardiology nurse disclosed cardiac-related SFs to 23 participants in a Finnish biobank [30]. In one large US-based biobank, a team comprising genetics clinicians and nonclinical support staff disseminated 515 SFs first to participants’ primary care providers (PCP) either via electronic health record (EHR) deposition or by telephone. Seven to ten days after PCPs were notified, participants were alerted that a SF had been uploaded to their EHR and following this, had their SF disclosed by a nonclinical support staff person following a result-specific telephone script [33].
Disclosure session duration
Secondary findings disclosure sessions for a total of 34 SF recipients were described as taking about one hour in three articles [4, 32, 38] with a range of 36–90 minutes.
Disclosure tools
A range of tools and materials used as part of SF disclosure were reported for 597 SF recipients across seven articles [4, 30, 32,33,34, 38, 44]. Summary letters provided to participants were mentioned in all but one of these articles [34]. Letters that SF recipients could use to facilitate family communication were mentioned in three articles, although it is unclear if these communication aids were uniformly provided [33, 38, 44]. The large biobank study that used EHR-mediated result return described at least seven distinct communication tools ranging from patient-facing disease-specific fact sheets to specific education modules targeting providers [33].
Disclosure method
Participants in a large US biobank (n = 515) were reported to have received their results initially by telephone yet had the option to meet in person or by telemedicine with a genetic counselor [33]. A Finnish biobank followed a similar procedure for their participants; notification letters were sent to 23 individuals with cardiac-related SF and then a cardiology nurse followed up with each individual by phone [30]. In ten additional articles describing how SF were disclosed to 73 individuals or families, disclosures were most often made in person during a clinic visit although several articles reported that some disclosures were made by telephone [4, 25, 27, 31, 32, 34, 36, 38, 43,44,45].
Provision of recommendations
Twenty-two articles describing SF disclosure to 646 SF recipients explicitly mentioned that SF recipients were provided with recommendations for management and/or referrals for additional care related to their SF [1, 4, 21, 22, 24,25,26,27,28, 30,31,32,33,34, 36,37,38, 40, 42, 44,45,46]. Twelve articles described recommending that SF recipients notify their families about their results so that relatives could consider cascade testing [22, 24, 25, 27, 28, 31,32,33, 36,37,38]. Several single family reports described how cascade testing of relatives was facilitated by the disclosing providers or team [25, 27, 37, 45].
Outcomes
Psychological impact
The psychological impact of receiving a SF was reported in 12 articles describing a total of 66 unique SF recipients [1, 4, 22, 23, 25, 30, 32, 36, 37, 40, 43, 44]. In these articles, the time from SF disclosure to assessment ranged between three months and four years. Reported emotions in these individuals included sadness, relief, empowerment, and surprise. In five articles describing psychological outcomes in 61 SF recipients, participants were specifically asked to describe feelings of regret after SF receipt and regret was only described by two participants [1, 4, 37, 43, 44]. In one article describing two SF recipients, a depression questionnaire (PHQ-9) and the Beck Anxiety Inventory were administered before and after SF disclosure to one participant; the difference between the scores at the two timepoints was reportedly not clinically significant [36]. Two other articles described studies assessing psychological impact both through questionnaires and via qualitative interviews in recipients of a variety of genomic findings that included SF as defined in this review, primary variants, and other actionable variants as defined by the study team and only aggregate results were reported in these. In one of these, most participants had positive or neutral feelings about their results [32] and in the other, overall low levels of distress were observed [43]. One SF recipient was described as not remembering their result [40].
Family communication
Nine articles described how a total of 52 SF recipients communicated their results with their relatives [1, 4, 22, 30, 36, 41, 44]. None of these included detailed information on family structure or pedigrees, limiting further analysis (e.g., what percentage of family members were notified). Two additional articles reported aggregated family communication of a variety of genomic results (including SF); in one, 93% of participants had shared their results [32] and the other reported that a majority of participants communicated with family [43]. Of their biologic relatives, SF recipients reported communicating their results to first-degree relatives most often although several reports of notifications to more distantly related relatives were reported (communication with 11 siblings, 16 parents, 11 children, and 7 more distant relatives was reported in the included articles). All four participants in one study described notifying their family members as a complex process due to their relatives’ personalities and the dynamics of their family [44]. In one instance, the index recipient of a BRCA1 SF learned that a relative had undergone clinical testing that identified the same variant, yet that relative had reportedly withheld this information from other family members to avoid causing concern [22].
Cascade testing
Twelve articles described subsequent cascade testing of at least one relative of 27 index SF recipients [1, 21, 23, 27, 36,37,38,39, 41, 44]. Several studies described cascade testing to determine the parent of origin of the SF [21, 23, 37, 39]. A total of 16 parents, 11 siblings, 11 children, and 7 second-degree relatives of SF recipients were reported as having undergone cascade testing in these articles, of these, 26 reportedly shared the SF recipient’s variant. Two articles listed one SF recipient each who had declined to pursue cascade testing of relatives facilitated by the disclosing providers [27, 36] and in a third, 11 of 18 SF recipients stated that none of their relatives had pursued testing to their knowledge when interviewed 6–18 months after SF disclosure [1].
Disease-specific evaluations and ongoing surveillance
At least one evaluation or consultation relevant to the SF was described in 144 index recipients in 17 articles [1, 4, 25, 27, 29, 31, 32, 34, 36,37,38,39, 41, 44, 46]. Nine of these articles described one evaluation, test, or consultation with a specialist in just one recipient of a SF [25, 34, 36,37,38,39, 41, 42, 46]. In two articles, 16 SF recipients underwent disease-specific evaluations, consultations, or both for cardiac conditions conducted by the research group as part of the SF disclosure process [32, 44]. Two articles described health-care actions in the year following SF disclosure in larger cohorts of SF recipients. One article used retrospective case reporting by clinicians familiar with the health-care actions of 66 SF recipients; 62 of these individuals had undergone at least one disease-specific evaluation [1]. Another article described the health-care actions of 48 women who had received BRCA1/2 SF as part of a biobank. In the year following return of the SF, 23 had undergone either a mammogram or breast magnetic resonance image (MRI) and 28 had met with a genetic counselor to discuss their results [29]. Four articles described ongoing surveillance associated with SF receipt for a total of six individuals. One SF recipient was described as undergoing regular cardiac screenings [25]; the remaining SF recipients were described as receiving ongoing imaging and other surveillance for cancer [21, 41, 46].
Provider communication
Five of the included articles described 36 SF recipients’ communication with a variety of health-care providers [1, 4, 25, 32, 43]. Of the 18 participants interviewed after SF disclosure in one study, seven reported informing their primary care provider and six reported discussing their results with a specialist [1]. Similarly, 15 of 18 recipients of a SF had shared their results with a health-care provider up to four years following SF disclosure in another study [32]; seven of 13 recipients shared their results with their primary care provider when interviewed four months after disclosure [4].
Treatment and prophylaxis
Sixteen individuals were described in eight articles as having undergone either risk-reducing prophylactic surgery or treatment for a manifestation of disease detected as a result of evaluations following SF disclosure [1, 4, 21, 26, 29, 38, 41, 46]. Medications were altered to avoid iatrogenic QT interval prolongation for one individual with an SCN5A variant [38]. One individual started HMG-CoA reductase inhibitor and a modified diet after return of an LDLR variant [21]. Two individuals with RET variants had thyroidectomies; one of these was described as prophylactic [21] and the other was done after a thyroid ultrasound identified nodules shown to be medullary thyroid carcinoma upon biopsy [26]. Another individual with an SDHB variant had a hemithyroidectomy after a biopsy showed early-stage papillary thyroid cancer [41]. Eleven women with BRCA1/2 SF were described as having undergone mastectomies and/or oophorectomies as part of cancer treatment or as risk-reducing prophylaxis [1, 4, 29, 38, 46].
Cost
Of the seven articles that described costs [1, 21, 29, 30, 38, 39, 46] one described sequencing costs only [21]. Another reported a fixed cost of $92,249.31 to start and run the pilot study through which participants were sequenced [38]. One Canadian study reported that the average cost to return genomic findings (including primary variants) was estimated at $750 per variant when a genetic counselor communicated the variant and $560 per variant when results were made available to the referring physician [39] and a Finnish study described the cost of returning heart disease related SF to biobank participants as the “…normal outpatient clinic fee ~27 EUR.” [30] In one article, median estimated follow-up costs ranged from $626 to $773 per variant disclosed to a total of 12 participants in a study which returned many different types of genomic variants; only one participant received a qualifying SF according to our definition [46]. Two articles reported costs associated with SF disclosure in a more specific way. One study calculated average costs of evaluations typically recommended for each gene–condition pair reported in their cohort and compared these calculations with observed health-care actions for 74 SF recipients (eight of which had been previously been reported by [32]) in the year immediately following disclosure. The average estimated cost of recommended care associated with SF was $421; the average observed cost was $128 [1]. These authors also estimated an average cost in provider wages of $31 associated with return of 48 SF. Only one study compared the pre- and postdisclosure health-care costs in a cohort of 48 recipients of BRCA1/2 SF and found that average difference in costs per patient was $2,578.00 (p = 0.76 per Wilcoxon signed-rank sum test) [29].
DISCUSSION
With this review, we synthesized and evaluated the available literature reporting how certain medically actionable genomic SFs are communicated and what outcomes are described after their receipt. The included articles described a wide spectrum of settings in which SFs became available, variation in disclosure practices, and disparate descriptions and ascertainment of postreceipt outcomes. These variations limited our ability to compare findings and use these results to optimize the efficiency and effectiveness of SF return. Outcome data were available for only a third of the SF recipients reported in the included articles and our review found that the evidence regarding outcomes associated with SF disclosure is weak. Eight years after the release of the first set of ACMG guidelines, the literature provided limited insight into how these guidelines have been implemented to achieve precision health-care outcomes for SF recipients and their families and suggests a number of areas for future research.
While almost all (646/709; 91%) of the SF recipients described were reported to have received recommendations for variant-specific medical care, the included literature provided more insight into disclosure practices than outcomes (Fig. 2). Having undergone at least one SF-specific evaluation, test, or consultation was the most frequently reported outcome in the included articles. Our review identified such outcomes in only 20% (144/709) of SF recipients but it is difficult to determine if that low fraction is representative of actual absence of such evaluations, or missing data. There was also a dearth of reports indicating that SF recipients were engaged in ongoing surveillance related to their finding; only six such descriptions were present in the included literature, although again, this could be attributable to missing data.
Three of the 13 included articles that focused on SF recipients were case reports describing a single individual or family experiences [25, 26, 37]. Seven articles reported findings from studies specifically designed to systematically relate SF disclosure to outcomes such as health-care use, cascade testing of relatives, and family communication; the 186 SF recipients described in these articles were all participants in research studies or biobanks employing sequencing to accomplish a variety of aims [1, 4, 29,30,31,32, 43]. None of these reported data on all seven of the outcomes we analyzed, and ascertainment methods used to report on each outcome varied among studies. In the five articles where the extent to which SF recipients underwent at least one evaluation or consultation related to their finding was assessed [1, 4, 29, 31, 32], the highest adherence rate (62/66; 94%) was reported by Hart et al. [1]. Adherence rates in the remaining four articles were 55% [32] to 72% [31]. However, more than one test or assessment is recommended as part of the evaluation of SF recipients for a clinicomolecular diagnosis [3] for nearly every disorder associated with ACMG SFTM. Data on the fraction of SF recipients who received thorough evaluations for a clinicomolecular diagnosis and continued adherence to a specific surveillance regimen were largely absent from the included literature. The small number of SF recipients reported and lack of detailed data limited our ability to draw conclusions about screening recommendation adherence. The paucity of outcome data in the included literature suggests that this is important area for future research.
The included literature emphasized the experiences of SF recipients who had research sequencing over those who received SF from clinical diagnostic testing. Of the 709 SF recipients reported in the included articles, only 21 underwent clinical sequencing [21,22,23,24,25,26]. While this might be expected given the ease of recruitment from existing studies and the fact that any individual clinical genetics practice will encounter SF in few patients, SF disclosure and outcomes may have distinct attributes in the clinical versus research settings. For example, those who receive SF in the clinical setting are likely to have a primary health condition that prompted the sequencing and may affect their perception of the relative importance of a SF in their overall health, and thus adherence to health behavior recommendations. The few articles that presented data from clinical sequencing may provide helpful insights but together only represent the experiences of a handful of the SF recipients reported here. We conclude that more data on SF from the clinical sequencing setting are urgently needed.
We applied the ACMG definition of clinical SF to our review because while they are clinical guidelines, numerous research initiatives apparently relied on this definition when returning results to their participants. However, secondary findings were defined heterogeneously across the included literature. In some articles, the terms “incidental” or “secondary” were used to describe genomic variants that were clearly associated with the sequenced individual’s presenting phenotype. For example, one study described “secondary variants” in both individuals who had reported a previous clinical diagnosis consistent with the finding and in individuals who would have met “criteria for genetic consultation and testing via standard clinical guidelines.” [34] Several additional studies employing tumor-normal sequencing used the terms as “incidental,” [28] or “secondary,” [31] when describing germline variants conferring increased cancer risk in cohorts of individuals with cancers clearly associated with the reported variants (e.g., germline BRCA1/2 variants found in women with breast and ovarian cancer). These variants were not included in our analyses because we considered them to be primary findings from indicated testing in affected individuals and not opportunistic screening. As mentioned, many research sequencing initiatives custom-select variants to return to their participants and in many cases these comprised those recommended by the ACMG along with several others (e.g., [32, 41, 43], Rego et al., 2018, Wynn et al., 2018). One article investigating the impact of receiving BRCA1/2 variants in biobank participants performed specific subanalyses on participants who had been unaware of their BRCA1/2 positive status; of the total cohort of 59 BRCA1/2 recipients reported in that article, only the 48 recipients who had been previously unaware of their status were used for that analysis [29]. Had these authors not reported these subanalyses, it would have been impossible to report cost data on true SF recipients. The inconsistency in how SF were labeled and defined across the included studies was itself an unexpected and notable finding of our review as consistent and broadly used definitions of a phenomenon are required prior to investigations into its nature. We conclude that clearer and stricter definitions of SF are needed and should be uniformly implemented in SF research to enhance generalizability of the findings.
Family communication and cascade testing of relatives has been known to be challenging in families seeking genetic testing for a primary indication [47,48,49,50,51,52]. Family communication and cascade testing were outcomes reported in few SF recipients described in the included literature (n = 52; 7% and n = 27, 4%, respectively). The limited available data do not permit conclusions to be drawn regarding the extent to which the experience of SF recipients paralleled that of families seeking indicated care. A similar paucity of data regarding SF recipient communication with health-care providers was demonstrated by our analyses. While primary care providers are reported to feel poorly equipped to incorporate genomic data into their practice [53, 54], the extent to which SF recipients in the reported literature perceived this is unknown.
The genomics community has engaged in a robust debate regarding the ethics implications and possible negative psychological impact of returning individual genomic findings with varying degrees of medical actionability [8, 10, 11, 15, 55]. While the data regarding psychological impact of SF receipt in the included articles were limited, we did not find evidence to support the hypothesis of significant psychological harm in SF recipients.
Stakeholder preferences on the return of SF and the ethics of SF was beyond the scope of our review. Sixteen times more articles were excluded for one of these two reasons than were included in our systematic analysis. A shift in focus away from some of the topics prioritized in the debate about returning genomic findings toward understanding the implementation of the ACMG guidelines is needed, especially as the guidelines will inevitably evolve.
Our review included two articles in which the cost associated with SF disclosure and medical follow-up was specifically assessed [1, 29]. Both of these articles concluded that the financial impacts associated with SF receipt in the one-year period following SF disclosure were lower than estimated or insignificant. These studies may be criticized for a lack of formal cost analysis rigor. Additional investigations following large cohorts of SF recipients recruited from research studies and those who learned of their SF from clinical sequencing are needed before the financial implications of SF disclosure can be better understood and robust cost–benefit analyses can be conducted.
Limitations
The variability in reporting of disclosure practices and outcomes measures limited our ability to analyze these elements of SF return and may have contributed to gaps in our analyses. Several of the included articles were case reports/case series reporting anecdotal data and none of the included articles reported on all of the elements we set out to analyze. While we designed our search terms to be inclusive, it is possible that articles describing the constructs of interest to this review were not captured by our search. Our definition of SF was challenging to apply to articles that described SF using terminology that did not distinguish primary from secondary genomic findings or those that described many genomic findings beyond those recommended for disclosure by the ACMG as medically actionable. In some instances, we had to work with aggregated data and these factors may have also led us to exclude articles that should have been included. The ACMG guidelines were intended to be implemented in the United States and seven of the articles included in our review originated in other countries where roles of personnel involved in SF disclosure, familial and societal norms, and access to health care can be quite different. We did not include articles describing disclosure practices or outcomes associated with receipt of negative SF.
Clinical and research implications
Our review suggests some preliminary conclusions and highlights several priorities for future research (Table 4). When this was assessed, a majority of SF recipients were described as undergoing at least one indicated evaluation related to their finding. While this suggests that SF recipients may receive health care consistent with a precision medicine approach, data suggesting a multidomain workup incorporating medical and family history, physical examinations, and thorough diagnostic testing to guide management are largely absent, as are investigations into, and reports of, determinants influencing this outcome. The individual, interpersonal, and systemic factors affecting outcomes associated with SF disclosure remain unknown. The experiences of SF recipients sequenced as part of research initiatives were overrepresented in our review. As sequencing becomes more widely used, understanding clinical SF disclosure practices and outcomes is an urgent priority because research participants and nonresearch patients may manifest important differences in how SF are received and managed. Finally, a cohesive and consistent definition of what constitutes a secondary genomic finding in the context of opportunistic screening is essential as there are clear inconsistencies and ambiguities in how SF are defined and considered.
Data availability
The submitted manuscript (including supplemental information) includes all data analyzed during this review.
References
Hart MR, Biesecker BB, Blout CL, Christensen KD, Amendola LM, Bergstrom KL, et al. Secondary findings from clinical genomic sequencing: prevalence, patient perspectives, family history assessment, and health-care costs from a multisite study. Genet Med. 2019;21:1100–1110.
Johnston JJ, Rubinstein WS, Facio FM, Ng D, Singh LN, Teer JK, et al. Secondary variants in individuals undergoing exome sequencing: screening of 572 individuals identifies high-penetrance mutations in cancer-susceptibility genes. Am J Hum Genet. 2012;91:97–108.
Katz AE, Nussbaum RL, Solomon BD, Rehm HL, Williams MS, Biesecker LG. Management of secondary genomic findings. Am J Hum Genet. 2020;107:3–14.
Sapp JC, Johnston JJ, Driscoll K, Heidlebaugh AR, Miren Sagardia A, Dogbe DN, et al. Evaluation of recipients of positive and negative secondary findings evaluations in a hybrid CLIA-research sequencing pilot. Am J Hum Genet. 2018;103:358–366.
Green RC, Berg JS, Grody WW, Kalia SS, Korf BR, Martin CL, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15:565–574.
Kalia SS, Adelman K, Bale SJ, Chung WK, Eng C, Evans JP, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med. 2017;19:249–255.
Miller DT, Lee K, Chung WK, Gordon AS, Herman GE, Klein TE, et al. ACMG SF v3.0 list for reporting of secondary findings in clinical exome and genome sequencing: a policy statement of the American College of Medical Genetics and Genomics (ACMG). Genet Med. 2021 May 20; https://doi.org/10.1038/s41436-021-01172-3. Online ahead of print.
Christenhusz GM, Devriendt K, Dierickx K. To tell or not to tell? A systematic review of ethical reflections on incidental findings arising in genetics contexts. Eur J Hum Genet. 2013;21:248–255.
Jackson L, Goldsmith L, O’Connor A, Skirton H. Incidental findings in genetic research and clinical diagnostic tests: a systematic review. Am J Med Genet A. 2012;158a:3159–3167.
Mackley MP, Fletcher B, Parker M, Watkins H, Ormondroyd E. Stakeholder views on secondary findings in whole-genome and whole-exome sequencing: a systematic review of quantitative and qualitative studies. Genet Med. 2017;19:283–293.
Ormond KE, O’Daniel JM, Kalia SS. Secondary findings: How did we get here, and where are we going? J Genet Couns. 2019;28:326–333.
Bertier G, Hétu M, Joly Y. Unsolved challenges of clinical whole-exome sequencing: a systematic literature review of end-users’ views. BMC Med Genomics. 2016;9:52.
de Wert G, Dondorp W, Clarke A, Dequeker E, Cordier C, Deans Z, et al. Opportunistic genomic screening. Recommendations of the European Society of Human Genetics. Eur J Hum Genet. 2021;29:365–377.
Delanne J, Nambot S, Chassagne A, Putois O, Pelissier A, Peyron C, et al. Secondary findings from whole-exome/genome sequencing evaluating stakeholder perspectives. A review of the literature. Eur J Med Genet. 2019;62:103529.
Salari P, Larijani B. Ethical issues surrounding personalized medicine: a literature review. Acta Med Iran. 2017;55:209–217.
All of Us Research Program I, Denny JC, Rutter JL, Goldstein DB, Philippakis A, Smoller JW, et al. The “All of Us” Research Program. N Engl J Med. 2019;381:668–676.
Kaye J, Hurles M, Griffin H, Grewal J, Bobrow M, Timpson N, et al. Managing clinically significant findings in research: the UK10K example. Eur J Hum Genet. 2014;22:1100–1104.
Moher D, Liberati A, Tetzlaff J, Altman DG; PRISMA Group. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: the PRISMA statement. Ann Intern Med. 2009;151:264–269.
Booth A, Papaioannou D, Sutton A. Systematic approaches to a sucessful literature review. Thousand Oaks, CA: SAGE publications; 2012.
Gale NK, Heath G, Cameron E, Rashid S, Redwood S. Using the framework method for the analysis of qualitative data in multi-disciplinary health research. BMC Med Res Methodol. 2013;13:117.
Baldridge D, Heeley J, Vineyard M, Manwaring L, Toler TL, Fassi E, et al. The Exome Clinic and the role of medical genetics expertise in the interpretation of exome sequencing results. Genet Med. 2017;19:1040–1048.
Basel D, McCarrier J. Ending a diagnostic odyssey: family education, counseling, and response to eventual diagnosis. Pediatr Clin North Am. 2017;64:265–272.
Catenacci DV, Amico AL, Nielsen SM, Geynisman DM, Rambo B, Carey GB, et al. Tumor genome analysis includes germline genome: are we ready for surprises? Int J Cancer. 2015;136:1559–1567.
Chirita-Emandi A, Andreescu N, Zimbru CG, Tutac P, Arghirescu S, Serban M, et al. Challenges in reporting pathogenic/potentially pathogenic variants in 94 cancer predisposing genes - in pediatric patients screened with NGS panels. Sci Rep. 2020;10:223.
Helm BM, Langley K, Spangler BB, Schrier, Vergano SA. Military health care dilemmas and genetic discrimination: a family’s experience with whole exome sequencing. Narrat Inq Bioeth. 2015;5:179–186.
Pendrick DM, Oberg JA, Hsiao SJ, Chung WK, Koval C, Sireci A, et al. Identification of a secondary RET mutation in a pediatric patient with relapsed acute myeloid leukemia leads to the diagnosis and treatment of asymptomatic metastatic medullary thyroid cancer in a parent: a case for sequencing the germline. Cold Spring Harb Mol Case Stud. 2019;5:a003889.
Leppig KA, Thiese HA, Carrel D, Crosslin DR, Dorschner MO, Gordon AS, et al. Building a family network from genetic testing. Mol Genet Genomic Med. 2017;5:122–129.
You YN, Borras E, Chang K, Price BA, Mork M, Chang GJ, et al. Detection of pathogenic germline variants among patients with advanced colorectal cancer undergoing tumor genomic profiling for precision medicine. Dis Colon Rectum. 2019;62:429–437.
Hao J, Hassen D, Manickam K, Murray MF, Hartzel DN, Hu Y, et al. Healthcare utilization and costs after receiving a positive BRCA1/2 result from a genomic screening program. J Pers Med. 2020;10:7.
Haukkala A, Kujala E, Alha P, Salomaa V, Koskinen S, Swan H, et al. The return of unexpected research results in a biobank study and referral to health care for heritable long QT syndrome. Public Health Genomics. 2013;16:241–250.
Horiuchi Y, Matsubayashi H, Kiyozumi Y, Nishimura S, Higashigawa S, Kado N, et al. Disclosure of secondary findings in exome sequencing of 2480 Japanese cancer patients. Hum Genet. 2021;140:321–331.
Lewis KL, Hooker GW, Connors PD, Hyams TC, Wright MF, Caldwell S, et al. Participant use and communication of findings from exome sequencing: a mixed-methods study. Genet Med. 2016;18:577–583.
Schwartz MLB, McCormick CZ, Lazzeri AL, Lindbuchler DM, Hallquist M, Manickam K, et al. A model for genome-first care: returning secondary genomic findings to participants and their healthcare providers in a large research cohort. Am J Hum Genet. 2018;103:328–337.
Thompson ML, Finnila CR, Bowling KM, Brothers KB, Neu MB, Amaral MD, et al. Genomic sequencing identifies secondary findings in a cohort of parent study participants. Genet Med. 2018;20:1635–1643.
CSER Consortium. https://cser-consortium.org. Accessed 5 January 2021.
Amendola LM, Lautenbach D, Scollon S, Bernhardt B, Biswas S, East K, et al. Illustrative case studies in the return of exome and genome sequencing results. Per Med. 2015;12:283–295.
Mackley M, McGuire K, Taylor J, Watkins H, Ormondroyd E. From genotype to phenotype. Circ Genom Precis Med. 2018;11:e002316.
Nestor JG, Marasa M, Milo-Rasouly H, Groopman EE, Husain SA, Mohan S, et al. Pilot study of return of genetic results to patients in adult nephrology. Clin J Am Soc Nephrol. 2020;15:651–664.
Papaz T, Liston E, Zahavich L, Stavropoulos DJ, Jobling RK, Kim RH, et al. Return of genetic and genomic research findings: experience of a pediatric biorepository. BMC Med Genomics. 2019;12:173.
Rego S, Dagan-Rosenfeld O, Bivona SA, Snyder MP, Ormond KE. Much ado about nothing: a qualitative study of the experiences of an average-risk population receiving results of exome sequencing. J Genet Couns. 2019;28:428–437.
Rego S, Dagan-Rosenfeld O, Zhou W, Sailani MR, Limcaoco P, Colbert E, et al. High-frequency actionable pathogenic exome variants in an average-risk cohort. Cold Spring Harb Mol Case Stud. 2018;4:a003178.
Westphal DS, Leszinski GS, Rieger-Fackeldey E, Graf E, Weirich G, Meitinger T, et al. Lessons from exome sequencing in prenatally diagnosed heart defects: a basis for prenatal testing. Clin Genet. 2019;95:582–589.
Wynn J, Martinez J, Bulafka J, Duong J, Zhang Y, Chiuzan C, et al. Impact of receiving secondary results from genomic research: a 12-month longitudinal study. J Genet Couns. 2018;27:709–722.
Ormondroyd E, Harper AR, Thomson KL, Mackley MP, Martin J, Penkett CJ, et al. Secondary findings in inherited heart conditions: a genotype-first feasibility study to assess phenotype, behavioural and psychosocial outcomes. Eur J Hum Genet. 2020;28:1486–1496.
Lee K, Berg JS, Milko L, Crooks K, Lu M, Bizon C, et al. High diagnostic yield of whole exome sequencing in participants with retinal dystrophies in a clinical ophthalmology setting. Am J Ophthalmol. 2015;160:354–363.e359.
Dewey FE, Grove ME, Pan C, Goldstein BA, Bernstein JA, Chaib H, et al. Clinical interpretation and implications of whole-genome sequencing. JAMA. 2014;311:1035–1045.
Menko FH, Ter Stege JA, van der Kolk LE, Jeanson KN, Schats W, Moha DA, et al. The uptake of presymptomatic genetic testing in hereditary breast-ovarian cancer and Lynch syndrome: a systematic review of the literature and implications for clinical practice. Fam Cancer. 2019;18:127–135.
Nycum G, Avard D, Knoppers BM. Factors influencing intrafamilial communication of hereditary breast and ovarian cancer genetic information. Eur J Hum Genet. 2009;17:872–880.
Roberts MC, Dotson WD, DeVore CS, Bednar EM, Bowen DJ, Ganiats TG, et al. Delivery of cascade screening for hereditary conditions: a scoping review of the literature. Health Aff (Millwood). 2018;37:801–808.
Sharaf RN, Myer P, Stave CD, Diamond LC, Ladabaum U. Uptake of genetic testing by relatives of lynch syndrome probands: a systematic review. Clin Gastroenterol Hepatol. 2013;11:1093–1100.
Whyte S, Green A, McAllister M, Shipman H. Family communication in inherited cardiovascular conditions in Ireland. J Genet Couns. 2016;25:1317–1326.
Wiseman M, Dancyger C, Michie S. Communicating genetic risk information within families: a review. Fam Cancer. 2010;9:691–703.
Christensen KD, Vassy JL, Jamal L, Lehmann LS, Slashinski MJ, Perry DL, et al. Are physicians prepared for whole genome sequencing? A qualitative analysis. Clin Genet. 2016;89:228–234.
Garrison NA, Brothers KB, Goldenberg AJ, Lynch JA. Genomic contextualism: shifting the rhetoric of genetic exceptionalism. Am J Bioeth. 2019;19:51–63.
Lohn Z, Adam S, Birch PH, Friedman JM. Incidental findings from clinical genome-wide sequencing: a review. J Genet Couns. 2014;23:463–473.
Acknowledgements
This study was supported by National Institutes of Health (NIH) grant HG200387-07. The authors are grateful to Alexandra Gomes and Elaine Sullor at George Washington University for their assistance with preliminary search strategies, M. Anna Buser and Jennifer Johnston for their critical review of the manuscript, and Darryl Leja for graphics support.
Author Information:
Conceptualization: J.C.S, K.L.L., P.v.d.W., D.C. Data curation: J.C.S, F.M.F. Formal analysis: J.C.S, F.M.F., E.M. Funding acquisition: L.G.B. Investigation: J.C.S, F.M.F., D.C., E.M. Methodology: J.C.S, D.C. Project administration: J.C.S, L.G.B. Resources: L.G.B. Supervision: J.C.S., P.v.d.W., L.G.B. Validation: J.C.S., F.M.F., D.C., K.L.L. Writing—original draft: J.C.S, F.M.F. Writing—reviewing & editing: J.C.S, K.L.L. F.M.F., P.v.d.W., D.C., E.M., L.G.B.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
L.G.B. is an uncompensated member of the Illumina Medical Ethics Committee, receives in-kind research support from Merck and Novartis, and honoraria from Cold Spring Harbor Press. The other authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Sapp, J.C., Facio, F.M., Cooper, D. et al. A systematic literature review of disclosure practices and reported outcomes for medically actionable genomic secondary findings. Genet Med 23, 2260–2269 (2021). https://doi.org/10.1038/s41436-021-01295-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-021-01295-7
This article is cited by
-
Invited Commentary on “My Research Results: a program to facilitate return of clinically actionable genomic research findings” by Willis et al.
European Journal of Human Genetics (2022)