Abstract
Purpose
An efficient framework to identify disease-associated genes is needed to evaluate genomic data for both individuals with an unknown disease etiology and those undergoing genomic screening. Here, we propose a framework for gene selection used in genomic analyses, including applications limited to genes with strong or established evidence levels and applications including genes with less or emerging evidence of disease association.
Methods
We extracted genes with evidence for gene–disease association from the Human Gene Mutation Database, OMIM, and ClinVar to build a comprehensive gene list of 6,145 genes. Next, we applied stringent filters in conjunction with computationally curated evidence (DisGeNET) to create a restrictive list limited to 3,929 genes with stronger disease associations.
Results
When compared to manual gene curation efforts, including the Clinical Genome Resource, genes with strong or definitive disease associations are included in both gene lists at high percentages, while genes with limited evidence are largely removed. We further confirmed the utility of this approach in identifying pathogenic and likely pathogenic variants in 45 genomes.
Conclusion
Our approach efficiently creates highly sensitive gene lists for genomic applications, while remaining dynamic and updatable, enabling time savings in genomic applications.
Similar content being viewed by others
INTRODUCTION
As genome and exome sequencing become standard in clinical genetic testing for patients with unknown genetic etiology and in broad genomic screening for population precision health, an efficient framework to identify and capture all known disease-associated genes is needed. With the scope of analysis in these assays covering over 20,000 genes, it is challenging to rapidly determine which genes have evidence of clinical relevance. Hence, a well-defined “medical exome”, consisting of genes with sufficient levels of evidence to warrant review in a clinical assay, is needed to limit the interpretative burden of reviewing variants from all genes.
There have been efforts to establish highly curated lists of gene–disease associations (GDAs), but these are often small. Most notably, the Clinical Genome Resource (ClinGen) has established a robust framework to determine gene–disease validity through manual assessment of strength of evidence that is used within their multiple disease-specific expert panels and working groups.1 While these GDAs are well curated, the intense effort required has limited the breadth of genes currently annotated. On the other end of the spectrum, computational tools, such as DisGeNET, attempt to classify the GDAs of all genes by integrating multiple databases into a single GDA score.2,3,4 However, the accuracy and validity of this scoring system has not been assessed. Other efforts have taken the approach of crowd-sourcing and/or collating GDAs, such as Genomics England’s PanelApp and the recently launched Gene Curation Coalition (GenCC), which allow diagnostic gene panels to be shared, downloaded, and evaluated by the scientific community, though they may be limited by the interests and thoroughness of the submitters.5,6
Generating and maintaining up-to-date gene lists remains challenging since assessing all GDAs is prohibitively time-consuming and evidence supporting new and existing GDAs is continuously generated. Previously published projects from our group, BabySeq and MedSeq, required manual curation resulting in a list of 1,514 and 1,490 GDAs, respectively. In both projects, this was a labor-intensive and time-consuming process that is not easily replicated in an efficient manner.7,8 Therefore, a balance between efficiency and thoroughness is required to make the analysis of genomic data more feasible.
Here, we propose a framework that balances efficiency, robustness, and accuracy to create gene lists for genomic analyses that can be routinely updated with new genes as associations emerge from the literature. This approach generates two lists of disease-associated genes based on different levels of evidence (comprehensive and restrictive) to be used in genomic applications.
MATERIALS AND METHODS
Data sources used to generate the comprehensive and restrictive gene lists
Extensive databases of gene and/or variant associations, including the Human Gene Mutation Database (HGMD), ClinVar, OMIM, and DisGeNET, were used to identify genes with any reported GDA.2,3,4,9,10,11 Each data source was also parsed to identify, when applicable, the number of classified variants and their review date, publications, and gene identifiers (Supplemental Methods).
Data sources used for validation of comprehensive and restrictive gene lists
Data sources incorporated for gene list validation included (1) 1,490 GDAs evaluated in MedSeq,8 (2) 1,514 GDAs evaluated in BabySeq,7 (3) 1,212 gene curations in 995 genes captured by ClinGen as of 14 March 2021,1 (4) 4,884 GDAs in the Incidentalome and Mendeliome panel from PanelApp Australia (accessed 26 February 2021),5 (5) 6,378 GDAs in the Paediatric panel from Genomics England PanelApp (accessed 26 February 2021),5 and (6) 2,187 GDAs across five laboratories in GenCC (accessed 13 March 2021).6 Each data set included a list of GDAs and their strength of evidence. Classifications derived from each data set and how they map to an overall strength of evidence are provided in Table S1 and defined in Supplemental Methods.
Genome sequencing and analysis
Genome sequencing data were generated from 45 individuals undergoing non-indication-based genomic screening (Supplemental Methods) with >30X mean coverage and a minimum completeness of >95% of all bases at ≥15X. Variants were filtered to the comprehensive or restrictive gene lists to identify pathogenic (P) or likely pathogenic (LP) variants (Supplemental Methods). Only genes mapping to GRCh37 were analyzed (Table S2). Evidence for GDAs was manually curated and each GDA was assigned one the following categories: (1) definitive, (2) strong, (3) moderate, or (4) limited using ClinGen criteria for gene–disease association. Following gene and variant curation using 2015 American College of Medical Genetics and Genomics/Association for Molecular Pathology guidelines12 with ClinGen rule specifications, only P/LP variants in genes with a strong or definitive GDA were considered reportable.
RESULTS
Generation of comprehensive and restrictive gene lists
To build a comprehensive gene list for clinical genomic applications, we extracted all genes from extensive data sets meeting any of the following criteria: (1) ≥1 P/LP variant in ClinVar, excluding copy-number variants (CNVs) overlapping multiple genes, (2) ≥1 variant classified as pathogenic (disease-causing mutation; DM) in HGMD, or (3) listed in Morbid Map from OMIM, excluding susceptibility and nondisease genes (Fig. 1a). Following these filters, the comprehensive list included 6,145 genes that have been implicated in Mendelian disease. Of note, 3,825 genes were present in all three data sets, with HGMD contributing the most unique genes (Fig. 1b).
(a) Schematic of the criteria fulfilled at each stage of the gene filtration process. Genes with entries in ClinVar (11,234 genes), OMIM Morbid Map (8,087 genes), and the Human Gene Mutation Database (HGMD) (12,080 genes) were integrated to generate the comprehensive and restrictive gene lists. Filtration parameters for each stage are presented in the right panel. (b) Venn diagram of the comprehensive (left) and restrictive (right) gene lists, including the number of genes meeting criteria in the initial databases. DM disease-causing mutation, P/LP pathogenic, likely pathogenic.
For many genomic applications, restricting the analysis to genes with stronger disease associations is preferable to reduce the burden on the laboratory. Thus, we further limited the comprehensive list by applying criteria using the number of P/LP variants, the recency of interpretation, and computational predictions for GDAs from DisGeNET. Specifically, only genes fulfilling any of the following criteria were retained: (1) ≥4 P/LP variants in ClinVar evaluated within the last 6 years (2015 or more recently) by any submitter, (2) ≥1 two-star P/LP variant in ClinVar, (3) mitochondrial genes with ≥1 P/LP variant in ClinVar, (4) ≥4 DMs in HGMD with supporting publications within the last 6 years (2015 or more recently), (5) genes with a DisGeNET GDA score ≥0.7. To add more stringency, we filtered this intermediate list to remove genes with lower levels of evidence, only keeping genes that met at least one of the following criteria: (1) ≥1 DM in HGMD with a supporting publication within the last 2 years (2019 or more recently), (2) ≥1 P/LP variant with a last evaluated date in ClinVar within the last 2 years (2019 or more recently), or (3) genes with a DisGeNET GDA score ≥ 0.3. All mitochondrial genes in the intermediate list were also kept at this stage. After applying both sets of filters, a restrictive gene list of 3,929 genes remained, with 3,427 genes present in all original data sources (Fig. 1).
Comparing gene lists to previous curations
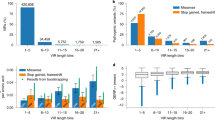
To determine the utility of the gene lists and specificity of the filtering strategy, we compared the comprehensive and restrictive lists to manual gene curations, including rigorous expert curations in ClinGen, manual gene assessments by an individual lab for BabySeq and MedSeq, crowdsourced approaches in PanelApp Australia and Genomics England PanelApp, and a consensus-based method from GenCC. When both lists were compared to the 995 genes from ClinGen, we observed that all definitive (655 genes) or strong (20 genes) gene–disease pairs in ClinGen were captured by both lists except for one definitive GDA missing from the restrictive list: the CD79B gene associated with agammaglobulinemia 6. This gene only had two P/LP variants in ClinVar and three DMs in HGMD. The latest ClinVar submission date was in 2007 and there were no publications after 2015 in HGMD (Table S2). Some gene–disease pairs with limited, disputed, refuted, or no evidence were removed from the comprehensive list (6.2%; 13/210), while many more were removed from the restrictive list (30%; 63/210) (Fig. 2a).
Gene lists were compared against GDA classifications provided by: (a) ClinGen, (b) MedSeq, (c) BabySeq, (d) consensus of Australian PanelApp (Incidentalome and Mendeliome panel) and Genomics England PanelApp (Pediatric Panel), and (e) consensus from GenCC. Numbers below the bar represent the number of genes included and numbers above the bar are the number of genes excluded in the respective list. Other: conflicting, refuted, disputed, no reported evidence, trait, pharmacogenomic association, only claim is from genome-wide association study (GWAS), and does not meet criteria. C Comprehensive Gene List, R Restrictive Gene List. aCD79B, bRPS15, cSMOC2.
When comparing the gene lists to the more rapid assessments of genes in MedSeq or BabySeq,7,8 we observed that all definitive or strong gene–disease pairs classified in both studies (603 genes and 951 genes, respectively) were captured by both lists, except for the strong RPS15 association with Diamond–Blackfan anemia curated in BabySeq that was not included in either gene list. The GDA between RPS15 and Diamond–Blackfan anemia was reassessed by the BabySeq team and downgraded to limited due to lack of supporting evidence. The comprehensive and restrictive gene lists also removed 51% (347/680) and 76.3% (519/680), respectively, of genes with insufficient or other classifications in MedSeq (Fig. 2b) and 6.2% (13/211) and 29.4% (76/211), respectively, of genes with limited or other classifications in BabySeq (Fig. 2c).
Additional analyses were performed using (1) a consensus interpretation from the largest panels of PanelApp Australia and Genomics England PanelApp and (2) a consensus GDA from GenCC. For the PanelApp analysis, most green-rated genes (1,956 genes) were captured by both lists, except for 41 genes (2.1%) removed from the restrictive list. The relatively high number of green-rated genes excluded from PanelApp in our restrictive list is expected as PanelApp is primarily focused on gene panels for in-depth diagnostic analysis and have not necessarily undergone extensive GDAs using rigorous criteria, such as is used in ClinGen. The gene lists also removed 12.9% (9/70) and 68.6% (48/70) of red genes from the comprehensive list and restrictive list, respectively (Fig. 2d). In the GenCC comparison, all definitive/strong genes were captured in both lists, except for SMOC2, associated with dentin dysplasia type I, that was missing from the restrictive list (Fig. 2e). This gene only had two DMs with the most recent publication in 2013 and 3 P/LP variants in ClinVar with the most recent evaluation date in 2017.
Genome sequencing results using different gene lists
To determine the performance of the gene lists in practice, genomic data from 45 individuals were screened for reportable variants using both the comprehensive and restrictive gene lists. Following variant filtration for putative P/LP variants, a total of 1,287 variants were identified in the comprehensive list, while only 1,096 were present in the restrictive list, a removal of 191 variants (15%; Fig. S1A). Per individual, this equated to an average of 29 (min = 14; max = 43) and 24 (min = 12; max = 35) variants in the comprehensive and restrictive lists, respectively. While 58% (402/696) of the genes in the comprehensive list met strong or definitive disease association after manual review, this ratio increased to 73% in the restrictive list (402/551). After variant assessment, all reportable variants from the comprehensive list—defined as P/LP associated to a strong or definitive GDA—were also identified in the restrictive list (an average of 3 variants per individual; min = 0; max = 7) (Fig. S1B).
DISCUSSION
Part of an effective and efficient strategy for exome and genome analyses includes defining an appropriate list of genes to interrogate for pathogenic variants. All genes with evidence for a disease association are needed for expanded analyses. However, in different contexts, the level of evidence required for the GDA may vary. For instance, genes with less or emerging evidence of disease association may be useful in settings where additional familial studies can help determine the likelihood of the gene’s responsibility for the individual’s disease. However, lists including limited evidence genes will have less utility in the context of genomic screening where the asymptomatic individual will not contribute evidence to the GDA and there is no or very limited utility of returning the result.
Here, we provide a framework that utilizes available databases to efficiently generate both a comprehensive (6,145 genes) and a restrictive list (3,929 genes) of disease-associated genes (Fig. 1; Table S1). Compared to ClinGen expert panels, the restrictive gene list excluded 30% of genes with lower levels of evidence, while maintaining all strong or definitively associated genes, aside from one gene with older and borderline evidence (Fig. 2). Additionally, applying the restrictive gene list to 45 genomes captured all reportable variants, while reducing the number of variants needed to be reviewed by 15%.
Further refinements to this approach can help further reduce the burden of genomic analyses, including utilizing more variant level information in the approach, such as handling variants with discordant classifications and variants whose population frequencies suggest they are too common to be associated with Mendelian disease. However, our current approach is easily implemented and updatable, shows high performance when compared to manually curated data sets, and can provide increased efficiency as genomic applications become more routine.
Data availability
The gene lists and data used to develop the lists can be found at https://Broad.io/genelist.
References
Strande, N. T. et al. Evaluating the clinical validity of gene-disease associations: an evidence-based framework developed by the Clinical Genome Resource. Am. J. Hum. Genet. 100, 895–906 (2017).
Piñero, J. et al. DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes. Database (Oxford). 2015, bav028 (2015).
Piñero, J. et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 45(D1), D833–D839 (2017).
Piñero, J. et al. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 48(D1), D845–D855 (2020).
Martin, A. R. et al. PanelApp crowdsources expert knowledge to establish consensus diagnostic gene panels. Nat. Genet. 51, 1560–1565 (2019).
The Gene Curation Coalition. https://thegencc.org/ (2021).
Ceyhan-Birsoy, O. et al. A curated gene list for reporting results of newborn genomic sequencing. Genet. Med. 19, 809–818 (2017).
Machini, K. et al. Analyzing and reanalyzing the genome: findings from the MedSeq Project. Am. J. Hum. Genet. 105, 177–188 (2019).
Landrum, M. J. et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 46(D1), D1062–D1067 (2018).
OMIM. https://www.omim.org/ (2020).
Stenson, P. D. et al. The Human Gene Mutation Database (HGMD®): optimizing its use in a clinical diagnostic or research setting. Hum. Genet. 139, 1197–1207 (2020).
Richards, S. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405–424 (2015).
Acknowledgements
Funding support was partly provided by grant 5R01HL143295 from the National Institutes of Health/National Heart, Lung, and Blood Institute (L.L.d.l.V., C.L.B.Z., R.C.G., H.L.R., M.S.L.). The authors would like to thank the Gene Curation Coalition (GenCC) for generating curated content used in this project. GenCC’s curated content was obtained at www.thegencc.org (13 March 2021) and includes contributions from the following organizations: Invitae, Illumina, Myriad Women’s Health, Ambry Genetics, and TGMI/G2P.
Author information
Authors and Affiliations
Contributions
Conceptualization: M.S.L., K.M,. C.A.A.T., H.M.S. Data curation: W.Y., L.H. Formal analysis: L.L.d.l.V., W.Y. Funding acquisition: R.C.G. Investigation: L.L.d.l.V., K.M. Methodology: M.S.L., H.L.R. Project administration: C.L.B.Z. Software: W.Y., L.H. Supervision: M.S.L. Visualization: W.Y., L.L.d.l.V. Writing—original draft: L.L.d.l.V., M.S.L. Writing—review and editing: H.L.R., C.L.B.Z., R.C.G., C.A.A.T., H.M.S. All authors reviewed and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics declaration
This project has been reviewed and approved by the Mass General Brigham institutional review board (IRB). All individuals consented for clinical genomic screening and all individual data was de-identified.
Competing interests
M.S.L., L.L.d.l.V., and C.L.B.Z. report grants from the National Institutes of Health (NIH) during the conduct of the study. H.L.R. reports grants from NIH during the conduct of the study; she also reports personal fees from Genome Medical outside the submitted work. R.C.G. reports grants from NIH during the conduct of the study; he also reports personal fees from AIA, SavvySherpa, Verily, and Wamberg, all outside the submitted work. The other authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Lazo de la Vega, L., Yu, W., Machini, K. et al. A framework for automated gene selection in genomic applications. Genet Med 23, 1993–1997 (2021). https://doi.org/10.1038/s41436-021-01213-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-021-01213-x