Abstract
Purpose
Diseases caused by defects in mitochondrial DNA (mtDNA) maintenance machinery, leading to mtDNA deletions, form a specific group of disorders. However, mtDNA deletions also appear during aging, interfering with those resulting from mitochondrial disorders.
Methods
Here, using next-generation sequencing (NGS) data processed by eKLIPse and data mining, we established criteria distinguishing age-related mtDNA rearrangements from those due to mtDNA maintenance defects. MtDNA deletion profiles from muscle and urine patient samples carrying pathogenic variants in nuclear genes involved in mtDNA maintenance (n = 40) were compared with age-matched controls (n = 90). Seventeen additional patient samples were used to validate the data mining model.
Results
Overall, deletion number, heteroplasmy level, deletion locations, and the presence of repeats at deletion breakpoints were significantly different between patients and controls, especially in muscle samples. The deletion number was significantly relevant in adults, while breakpoint repeat lengths surrounding deletions were discriminant in young subjects.
Conclusion
Altogether, eKLIPse analysis is a powerful tool for measuring the accumulation of mtDNA deletions between patients of different ages, as well as in prioritizing novel variants in genes involved in mtDNA stability.
Similar content being viewed by others
INTRODUCTION
The human mitochondrial genome (mtDNA) is a double-stranded molecule, encoding 13 peptides involved in oxidative phosphorylation (OXPHOS), together with 2 ribosomal RNAs (rRNA) and 22 transfer RNAs (tRNA).1 Pathogenic variants can affect either protein coding genes2 or tRNA and rRNA coding genes.3,4,5,6 MtDNA can be altered by large deletions or duplications and depletion, all of which are common causes of mitochondrial disorders,7 in particular in individuals harboring pathogenic variants in proteins involved in mtDNA maintenance.8 The mutant load varies drastically among different tissues, from 100% mutant load, a condition known as homoplasmy occurring with a small number of variants of low penetrance, to the combination of mutant and wild-type copies, described as heteroplasmy, often leading to a threshold beyond which clinical symptoms appear.1,9
Next-generation sequencing (NGS) of the mitochondrial genome from muscle biopsy is considered as the gold standard procedure for diagnosing mitochondrial disorders related to mtDNA instability,10 while the data obtained from blood samples are less informative.11 Recently, sequencing data obtained from uroepithelial cell samples were reported to be sensitive enough without invasive procedures.11,12
MtDNA rearrangements accumulate physiologically with age in postmitotic tissues and in mtDNA maintenance disorders. They are mainly located in the major arc of the mtDNA,13 but in-depth characterization remains a challenge.14 In general, mtDNA deletion and duplication breakpoints are located near homologous repeat sequences, distinguishing rearrangements occurring in direct perfect repeats (type I deletion) from those occurring in imperfect repeats or sequences without similarities (type II deletion).15,16
To date, more than 20 genes have been implicated in the maintenance and replication machinery of the mitochondrial genome.6 Variants in POLG encoding the DNA polymerase are among the most common causes of accumulation of mtDNA multiple deletions, encompassing a large spectrum of symptoms.17,18 Nevertheless, identification of novel variants of unknown significance (VUS) in nuclear genes involved in mtDNA maintenance requires a molecular validation to confirm their pathogenicity.
In this work, we report an in-depth analysis of mtDNA sequences from patients with mitochondrial disease and controls, using NGS data processed through eKLIPse, leading to the identification of specific criteria distinguishing mtDNA rearrangements related to aging from those related to mtDNA maintenance defects.
MATERIALS AND METHODS
Patient samples
Full clinical and molecular descriptions of patients are summarized (Tables S1–S3). Total DNA were extracted from muscle, blood, and urine as described elsewhere.19
Retrospective study: derivation cohort
Skeletal muscles (n = 53) and uroepithelial cells (n = 37) from controls displayed normal mitochondrial enzyme assays and OXPHOS complex assembly. Similarly, muscle and uroepithelial control samples were screened for the absence of pathogenic variants in mtDNA maintenance genes.
In parallel, we analyzed skeletal muscle samples (n = 32) and uroepithelial cells (n = 9) from individuals with pathogenic variants in genes involved in mtDNA instability classified according to American College of Medical Genetics and Genomics/Association for Molecular Pathology (ACMG/AMP) criteria20 (Table S1).
Prospective study: validation cohort
Over a one-year period of clinical diagnosis, eKLIPse was used to look at the presence of mtDNA deletions in a series of 15 skeletal muscles and 2 uroepithelial samples from 17 patients (Table 2). For patients 41 to 54, mtDNA sequencing was performed as a firstline test, while for patients 55 to 57, mtDNA sequencing was performed after the initial identification of VUS in mtDNA maintenance genes.
Patient 55 (P55), a 23-year-old man, was followed for chronic progressive external ophthalmoplegia (CPEO); he was previously reported as patient 4.19 Analysis of a targeted gene panel identified two POLG variants (Table 2) classified as likely pathogenic and VUS. The mtDNA from muscle and uroepithelial cells was analyzed.
Patient 56 (P56) was a 46-year-old man suffering from CPEO and dysphagia since the age of 18. Family history disclosed numerous affected individuals over three generations. Molecular analysis identified a heterozygous variant in TWNK (Table 2), classified as likely pathogenic. The mtDNA from uroepithelial cells was analyzed.
Patient 57 (P57), a 63-year-old man, was followed for ptosis and CPEO from the age of 30. His 43-year-old daughter had the same disease diagnosed at similar age. Molecular analysis identified a TWNK heterozygous variant (Table 2) classified as VUS. The mtDNA from muscle obtained at age 53 was analyzed.
mtDNA next-generation sequencing and bioinformatic processing
The entire mtDNA was amplified in overlapping fragments using a combination of primers designed to avoid nuclear pseudogene amplification. Primer sequences and polymerase chain reaction (PCR) conditions are available in Supplementary methods. Ion Torrent® single-end sequencing (n = 115) and Illumina® paired-end sequencing (n = 16) were performed as described elsewhere.19 Sequencing data were processed through a previously described homemade pipeline21 including eKLIPse.19
Statistical analysis
Qualitative variables were compared using Fisher’s exact test, while quantitative variables were analyzed by the nonparametric Mann–Whitney test. When necessary, a Benjamini–Hochberg correction was applied. Multiple comparisons were performed using a one-way univariate analysis of variance (ANOVA) and the nonparametric Kruskal–Wallis test. Comparisons by pairs were performed using Steel–Dwass test. Values of p < 0.05 were considered statistically significant. Statistical analyses were performed using JMP 10.0.0 statistical software (SAS Institute, NC, USA). Based on univariate and multivariate results, a neural network approach was assessed using the predictor profile (JMP).
RESULTS
MtDNA deletion signatures of 40 patients carrying pathogenic variants in nuclear genes involved in mtDNA maintenance (Table S1) were compared to 53 controls, according to age, using muscle and urine samples. MtDNA deletions were identified in about two-thirds of the controls, either in muscle (n = 35, 66%) or uroepithelial cells (n = 24, 64.9%) (Table 1), with a total of 362 different mtDNA deletions identified: 217 deletions in skeletal muscles (60%) and 145 in uroepithelial cells (40%) (Table S2). Data from muscle and uroepithelial controls were separated into four groups, according to age (Fig. 1): group A: individuals under 15 years (muscle n = 6, urine n = 8), group B: individuals aged between 15 and 30 years (muscle n = 11, urine n = 10), group C: individuals aged between 30 and 50 years (muscle n = 16, urine n = 8) and group D: individuals aged over 50 years (muscle n = 20, urine n = 11).
Individuals were subdivided into four groups according to age. A Circos representative of one individual is shown for each group. N = number of patients/group. Selected parameters between patients compared to controls are represented as boxplots: deletion number, length of breakpoint repeats, heteroplasmy levels, maximum cumulative percentage of deletions. Statistical significances are *p < 0.05, **p < 0.01, ***p < 0.001.
Multiple mtDNA deletions accumulate in control skeletal muscle and uroepithelial cells during aging
In skeletal muscle, a significant age-related mtDNA deletion accumulation was identified (p = 0.031) with, on average, 0.8 (±1.0) deletions in group A, 1.3 (±1.8) in group B, 3.5 (±7.5) in group C, and 7.1 (±8.1) in group D, visualized as multiple red arcs in Circos representations (Fig. 1). Although the deletion number was not significantly different between groups before the age of 50, the accumulation of deletions became significant in individuals from group D, compared to other age groups (group A, p = 0.018; group B, p = 0.036; group C, p = 0.045) (Fig. S1).
Characteristics of deletion profiles were analyzed in controls over the age of 15 (groups B to D). The size of deletions did not vary significantly with age (p = 0.09), with mean deletion lengths of 6,142 bp, 5,598 bp, and 5,699 bp in groups B, C, and D respectively. Similarly, the cumulative percentage of deletions did not vary significantly between the different groups (p = 0.08), ranging from 0.06% to 1.54% to 1.21% in groups B, C, and D, respectively. Heteroplasmy levels only varied significantly with age (p < 0.0001), first increasing until the age of 50, then decreasing, with a mean of 0.03%, 0.29%, and 0.18% in groups B, C and D, respectively. The control region (m.16024 to m.576), was partially or completely absent in 18.9% of the deletions of controls (Table 1).
In uroepithelial cells, mtDNA deletions accumulate during aging, with on average, 0.9 (±1.1) deletions in group A, 2.5 (±3.1) in group B, 3.9 (±7.8) in group C, and 7.5 (±9.7) in group D, although without reaching significance (p = 0.12), with the exception of the comparison between groups A and D (p = 0.0257) (Fig. S2). The control region was removed partially or completely in 42.8% of samples (Table 1).
MtDNA deletion profiles in muscle, comparing patients with controls
MtDNA deletion profiles in muscle revealed a significantly higher number of deletions in patients carrying a mtDNA maintenance defect than in controls (Fig. S1; Table 1). All patients (n = 32) carried multiple mtDNA deletions, while only 66% of controls (n = 35/53, p < 0.001) did so. The deletion number was 20 times higher in patients (84.1 ± 91.5) than in controls (4.1 ± 6.9) (Table 1), even after subjects had been stratified according to age (Fig. 1) with 14.0 (±15.0) vs. 0.83 (±0.98) deletions in group A (p = 0.0391), 98.3 (±74.2) vs. 1.3 (±1.8) in group B (p = 0.008), 42.3 (±36.9) vs. 3.5 (±7.5) in group C (p = 0.001), and 109.8 (±104.9) vs. 7.1 (±8.1) in group D (p < 0.0001), respectively. A comparison of mtDNA deletion signatures in patients and controls disclosed that deletion sizes were not significantly different (p = 0.6, Table 1). Conversely, heteroplasmy levels were increased in patients (p < 0.001) regardless of age (Fig. 1), with an average heteroplasmy level of 0.25% (±0.55%) vs. 0.03% (±0.05%) in group B (p < 0.0001), 0.55% (±1.19%) vs. 0.29% (±0.62%) in group C (p = 0.0154), and 0.33%(±1.73%) vs. 0.17% (±0.51%) in group D (p = 0.0002), in patients and controls, respectively. Similarly, the maximum cumulative heteroplasmy levels were significantly higher in patients (23.21%), than in controls (0.99%) (Table 1; Fig. 1), values equal to 21.7% (±14.2%) in patients vs. 0.07% (±0.06%) in controls in group B (p = 0.028), 18.66% (±21.1%) vs. 1.54% (±2.1%) in group C (p = 0.013), and 28.6% (±26.9%) vs. 1.2% (±1.4%) in group D (p < 0.0001), respectively. In terms of breakpoints, we assessed the nucleotide sequence surrounding the deletion revealing that the locations of mtDNA deletions in patients were different from those of controls, with breakpoints occurring closer to perfect repeats in controls than in patients (4.4 bp vs. 2.9 bp, p < 0.001) (Table 1; Fig. 1), independently of age, with a repeat length of 4.5 bp (±3.5) vs. 2.0 bp (±2.5) in group B (p = 0.001), 4.9 bp (±4.2) vs. 2.6 bp (±2.6) in group C (p = 0.0008), and 4.3 bp (±3.9) vs. 3.1 bp (±3.1) in group D (p = 0.0018) in controls and patients, respectively. Moreover, in contrast to what was observed in controls, few deletions encompassed the entire control region in patients (12.9% vs 1.5%; Table 1).
mtDNA deletion profiles in uroepithelial cells and blood from patients and controls
Metric comparisons were only assessed between patients and controls without stratification by age due to the limited number of samples in each group (Table 1, Fig. S4). As in muscle, mtDNA deletions accumulated significantly in patient uroepithelial cells compared to controls. Indeed, deletions were present in all patient samples (n = 9), while only in 64.9% of controls (n = 24/37, p = 0.023), with an average number of 8.2 in patients and 3.9 in controls (Table 1; Fig. S2, S3). In contrast to muscle samples, mtDNA deletion sizes were significantly smaller in patients than in controls (4,959 bp vs. 6,553 bp), paralleling a lower heteroplasmy rate (0.17% vs. 0.25%), while repeat lengths at breakpoints were not significantly different between both groups (3.14 bp vs. 2.68 bp). Furthermore, the analysis revealed a higher level of mtDNA deletions encompassing partially or entirely the D-loop region in control individuals, compared to patients (42.8% vs 2.7%, p < 0.0001). In parallel, DNA samples extracted from blood of controls (n = 48) versus patients with mtDNA maintenance defects (n = 28) were analyzed and very few deletions were detected through eKLIPse.
Data mining and interpretation of mtDNA deletion profiles
A supervised learning algorithm model was developed using eKLIPse data regarding muscle, including the following parameters: patient age, deletion number, heteroplasmy level, mean deletion length, mean length of the repeats at breakpoints, deletion number encompassing the D-loop region, and the maximum cumulative heteroplasmy levels (Fig. 3a; Tables S2, S3). A total of 67 samples were selected for the learning algorithm, subdivided into 44 for the training test, and 23 for the validation test (Fig. 3a). Misclassification rates were lower than 10%, with 9.1% for the training and 8.6% for the validation sets.
Then, mtDNA deletion profiles from 17 patients were analyzed to assess eKLIPse performances in two situations frequently encountered in clinical diagnostic laboratories (Table 2): first, when a specific mtDNA deletion signature was identified (patients 41 to 54), and second, when questioning the pathogenicity of novel nuclear variants identified in genes involved in mtDNA maintenance (patients 55 to 57).
Due to the number and required combinations of parameters for analysis, a learning algorithm was modeled on mtDNA deletion signatures to decipher the individual status with a final probability score. For patients 41 to 54, mtDNA sequencing from muscle or uroepithelial samples was performed as a firstline test. Between 2 and 53 deletions per sample were detected (Table 2), with specific deletion patterns analyzed using a neural network approach (Fig. 3a). Based on the training test, a probability score above a threshold of 0.55 was considered significant in cases classified as patients. Between 0.45 and 0.55, the patient’s status was considered as undetermined, and below 0.45 as control. According to their prediction profiles, eight samples were classified as controls, two as undetermined (P44 and P51), and seven as patients carrying a mtDNA maintenance defect (P50, P52–57) (Table 2).
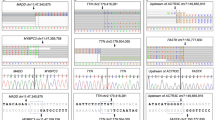
Interestingly, among individuals classified as patients, the 77-year-old patient P52 without a known family history was asymptomatic until age 70, with later a diagnosis of a progressive girdle muscular dystrophy and polyneuropathy, associated to COX negative fibers. EKLIPse analysis revealed a total of 25 different deletions in muscle, which was classified as a patient with a score of 1.0 according to the prediction profiler (Table 2), including the m.7398-13782del with a mutant load of 43.3%, and a maximum cumulated deletion rate of 51%, as shown in the Circos representation in Fig. 2a. However, by removing the m.7398-13782del deletion from the analysis, the remaining 24 deletions would have been classified as control by the prediction profiler with a final score of 0.303. In this case, the presence of the m.7398-13782del deletion in muscle was evidence of a pathological condition, whereas all other deletions were related to aging. Notably no deletion was found in patient uroepithelial cells.
Circos graphical plots show mtDNA deletions as red arcs with their intensity proportional to the mutant load. (a) In muscle from patient P52 carrying a sporadic mtDNA deletion with additional age-related mtDNA multiple deletions. (b) In uroepithelial cells from patient P54 carrying a heterozygous TWNK variant. (c) In muscle and uroepithelial cells from patient 55 carrying two POLG pathogenic variants. (d) In uroepithelial cells from patient P56, carrying a heterozygous TWNK variant. (e) In muscle from patient P57 carrying a TWNK variant.
For patients P44 and P51 classified as undetermined, biochemical assays on skeletal muscle (Table 2) revealed a normal respiratory chain in P44 and a complex I deficiency in P51. Targeted sequencing of genes involved in mtDNA maintenance was not informative.
In patients for whom a mtDNA maintenance defect was predicted, the molecular diagnosis disclosed POLG compound heterozygous variants in patient P53 (Table 2). For patient P54, a total of 53 deletions were identified in uroepithelial cells with a maximum cumulative deletion reaching 6% (Fig. 2b) compared to control cells from age-matched group D, with an average of 7.5 deletions (±9.7) and 2.1% of maximum cumulated deletions (±4.4). A TWNK variant was identified, classified as likely pathogenic, as already seen in patient P29 from the derivation cohort with a similar clinical phenotype.
Characterization of mtDNA multiple deletion profiles allows for the functional validation of novel variants in mtDNA maintenance nuclear genes
The 21-year-old patient P55 was suffering from ophthalmoplegia and dysphagia, carrying 2 POLG variants classified as VUS according to ACMG/AMP criteria (Table 2). Three hundred thirty-eight deletions were identified in muscle sample, with a maximum cumulative deletion level reaching 64% compared to an average of 3.5 (±7.5) deletions and maximum cumulative heteroplasmy of 0.06 (±0.06%) in the age-matched control group B (Fig. 2c). The predicted probability of presenting a mtDNA maintenance defect reached 99.8% according to patient age (Table 2). In uroepithelial cells, 27 deletions were identified (Table S3) with a percentage of maximum cumulative deletions reaching 17%, compared to an average of 2.5 (±2.9) deletions and cumulative heteroplasmy of 0.12% (±0.14%) in age-matched control group B. Together, these data supported a damaging effect of POLG variants reclassified as pathogenic and likely pathogenic.
Patient P56 had a single TWNK variant (Table 2) and mtDNA analysis from urothelial cells revealed a total of 13 deletions with a maximum cumulative deletion reaching 2.4% (Fig. 2d), significantly higher than the median 0.09% of age-matched control group C. None of the deletions encompassed the D-loop, and breakpoints were not located within perfect direct repeats. This abnormal mtDNA pattern strongly supported the damaging effect of the TWNK variant, reclassified as pathogenic class 5, according to ACMG/AMP.
Patient P57, affected with CPEO, was carrying a single TWNK variant. The analysis revealed 16 mtDNA deletions in muscle (Fig. 2e), none affecting the D-loop. According to the algorithm model, the probability of an mtDNA maintenance defect was estimated at 0.997 (Table 2), supporting the damaging effect of the TWNK variant, and was reclassified as likely pathogenic class 4.
DISCUSSION
Alterations of mitochondrial genome integrity cause a variety of clinical presentations, resulting from mtDNA depletion and multiple deletions and/or duplications.14,22 It has always been difficult to evaluate the presence of mtDNA deletions with respect to their contribution to disease or mtDNA rearrangements related to aging. To address this question, we developed eKLIPse, which is based on soft clipping and predicts genome breakpoints by realigning soft-clipped sequences that do not align to the mtDNA reference sequence.19 The detection of abnormal mtDNA profiles should then promote targeted nuclear gene panels including mtDNA maintenance genes or exome sequencing as described in the flowchart (Fig. 3b). With this tool in hand, it became mandatory to challenge its capabilities to analyze a large number of patient samples, in order to evaluate eKLIPse performances for the diagnosis of mtDNA instability disorders.
(a) Representation of the Multilayer Perceptron neural network for classifying patients and controls according to age, deletion number, deletions removing the D-loop, deletion length, percentage of deletions, sequence repeats, and maximum cumulative heteroplasmy. (b) Receiver operator characteristic (ROC) curves of the learning and validation phases. A data set of 67 muscle samples (35 controls and 32 patients) (Supplementary Table 2) was used to develop the machine learning approach. Data were randomly divided into the learning data set (44 samples) and the validation data set (23 samples). (c, d) Schematic goals of eKLIPse for the diagnosis of disorders related to mitochondrial genome instability. (c) Orientation of the diagnostic strategy. (d) Functional validation of variants of unknown significance (VUS) in genes involved in mtDNA maintenance.
To address this objective, we analyzed 53 and 32 muscles, and 37 and 9 urine samples, from controls and patients respectively. In all possible comparisons from the study, our results disclosed significant differences when comparing patient versus control mtDNA profiling, according to different age groups or tissue samples, i.e., muscles versus uroepithelial cells.
The comparison of mtDNA deletion profiles in controls and patients provided new criteria to better diagnose mitochondrial disease patients with mtDNA instability. The selection of mitochondrial parameters was highly dependent on patient age. The number of multiple deletions was much higher in adults compared to children, representing a biomarker for the diagnosis of mtDNA maintenance defects. The best signature was obtained in patient muscle showing an unambiguous profile of multiple deletions carrying pathogenic variants related to mtDNA maintenance according to different age groups, thus distinguishing mtDNA maintenance defects from age-related mtDNA deletions.
Additional parameters were identified as discriminating markers able to differentiate deletions due to aging from defects in the mtDNA maintenance. Interestingly enough, the number of deletions encompassing part or all of the D-loop region was significantly higher in control individuals compared to patients, as seen previously.23 It has previously been suggested that the origins of replication OH and OL were rarely deleted in cases of multiple deletions due to nuclear maintenance defects, while the origin of replication of the light strand was deleted in more than 20% of age-related mtDNA deletions.23 The presence of deletions encompassing the D-loop region or origins of replication are witnessing the reduced turnover rate of damaged mitochondria during the aging process.24
The absence of short repeat lengths surrounding the deletions in young patients compared to age-matched controls was a discriminating parameter. This was in line with the identification of type II breakpoints without direct repeats seen in children severely affected with mtDNA deletion syndrome, considered as a marker of severity.25
The maximum cumulative percentage of mtDNA deletions given by eKLIPse at specific nucleotide position or gene was greater in the muscle of patients compared to the age-matched controls. This was exemplified by the analysis of the 77-year-old patient P53 with late-onset disease whose first signs started at the age of 70, for whom we found evidence of a mixture of age-related deletions and a sporadic large-scale rearrangement in muscle. Late-onset clinical manifestations were probably linked to the synergistic effect of both deletion types highlighting the clinical usefulness of the determination of the maximum cumulative percentage of mtDNA deletions. This case illustrates the importance of detecting and quantifying age-related deletions in patients with mtDNA instability, which may precipitate or worsen the clinical phenotype with a different array of symptoms linked to mutant loads as already seen with large-scale mtDNA deletions.26
We evaluated in parallel the usefulness of analyzing urine sediment in patients to potentially avoid invasive muscle biopsies. The type of samples required to evaluate the presence of mtDNA rearrangements is an important issue.27,28 Except muscle tissue, which is considered as a gold standard, the type of sampling required to analyze a patient with mitochondrial disease has been a matter of debate for years, especially by opposing the use of DNA extracted from blood to other tissues, such as uroepithelial cells.11,12
It has been demonstrated previously that mutant loads of pathogenic variants in uroepithelial cells were comparable to muscle11,12 but appeared in our study not strictly similar for mtDNA rearrangements between both tissues. Indeed, uroepithelial cells also displayed the accumulation of multiple deletions to some extent, but the generated profile was not as strong compared to muscle as exemplified by patient P56 with 338 deletions identified in muscle and 27 deletions in uroepithelial cells. The deletion number in uroepithelial cells was still significantly higher when compared with control individuals, making it helpful for evaluating the molecular signature in patients with mitochondrial disease. Differences between muscle and urine samples may be due to the cellular heterogeneity of uroepithelial cells carrying varying levels of mtDNA rearrangements.29 This is in line with a recent study looking at the mutant load and the mtDNA copy number to evaluate disease burden in patients carrying the m.3243A>G common variant.30 Urine was considered as a useful tissue for variant detection, but with high variability of heteroplasmy levels due to differences in cellular composition and gender30 or compared to pathogenic variants, it may have relied on a specific deletion formation mechanism influencing the level of rearrangements.31
It has recently been proposed that urinary sediment cells might be useful for the detection of sporadic large-scale mtDNA deletions as an alternative to muscle biopsy.32 Similar to our study, the heteroplasmy level of single large-scale mtDNA deletions in nine PEO patients was lower in uroepithelial cells versus muscle, but the number of patients was too limited to be able to drive a final conclusion.32 More longitudinal studies are required to evaluate the usefulness of the detection and quantification of deletions in urine sediment.
Finally, to facilitate the interpretation of mtDNA deletion signatures, all selected parameters were assessed together. We used data mining and a learning algorithm to differentiate patients with mtDNA maintenance defects from the accumulation of deletions through aging.14 We have trained and validated our model through sequencing data obtained with a patient set carrying pathogenic variants of mtDNA maintenance defects and a control set. Although determining the exact sensitivity and specificity will require more data, this study further supports the potential for mtDNA deletion profiles through eKLIPse as a diagnostic test and functional evaluation of novel variants affecting nuclearly encoded mtDNA maintenance genes (Fig. 3b). We have, for example, been able to give precise molecular diagnoses for patients bearing VUS in POLG and TWNK genes, providing a final genetic diagnosis.
To date, there are no obvious correlations between phenotype and genotype through the accumulation of multiple deletions. Patients harboring mtDNA rearrangements frequently have a similar spectrum of clinical phenotypes including CPEO, ptosis, or myopathy to those seen in patients with pathogenic variants in tRNA genes.33 This is important information reflecting the consequences of the accumulation of multiple deletions, highlighting that most rearrangements commonly encompass one or more tRNAs affecting mitochondrial protein synthesis, and demonstrating that the cumulative percentage of mtDNA multiple deletions may have detrimental in vivo effects.34
So far, most studies have been undertaken on patient cohorts with sporadic large-scale mtDNA deletions.32 The determination of the exact size, location, and heteroplasmy level of deletions appeared to be of clinical importance.35 It was shown that larger deletions are often associated with early disease onset and more severe clinical symptoms; they also depend on the percentage and location of deletions that have been shown to distinguish between CPEO and Kearns–Sayre phenotypes.36 Another study suggested that increased levels of deletions but also lowered sequence homology flanking the deletion were correlated with more severe clinical phenotype and early onset.16 Grady et al. have indicated that disease progression and severity were correlated with heteroplasmy level, site, and size of single large-scale deletions.37
In addition, eKLIPse will likely be extremely useful for looking at the accumulation of somatic deletions. Somatic mtDNA deletions may be important in exacerbating the onset and progression of mtDNA diseases but also of age-related diseases.7 eKLIPse offers the potential for uncovering the spectrum of mtDNA rearrangements in somatic tissues as seen previously in subtantia nigra neurons.38
MtDNA deletion profiles generated by full mtDNA sequencing and eKLIPse analysis allowed for an unprecedented qualitative and quantitative characterization of patients with mtDNA maintenance defects, improving the molecular diagnosis in particular when facing the identification of VUS39 and then better management and genetic counseling of patients with mtDNA instability related disorders.
Data availability
EKLIPSe software is available through github (https://github.com/dooguypapua/eKLIPse). The diagnostic algorithm was designed using the commercially available JMP software package (JMP 10.0.0 statistical software from SAS Institute, NC, USA. All patient clinical and molecular data supporting the conclusions of this study are presented within the article and Supplementary Tables 1–3.
References
Wallace, D. C., Fan, W. & Procaccio, V. Mitochondrial energetics and therapeutics. Annu. Rev. Pathol. 5, 297–348 (2010).
Lott, M. T. et al. mtDNA variation and analysis using Mitomap and Mitomaster. Curr. Protoc. Bioinformatics. 44, 1 23 21–26 (2013).
Gorman, G. S. et al. Prevalence of nuclear and mitochondrial DNA mutations related to adult mitochondrial disease. Ann. Neurol. 77, 753–759 (2015).
Tang, S. et al. Transition to next generation analysis of the whole mitochondrial genome: a summary of molecular defects. Hum. Mutat. 34, 882–893 (2013).
Elson, J. L. et al. The presence of highly disruptive 16S rRNA mutations in clinical samples indicates a wider role for mutations of the mitochondrial ribosome in human disease. Mitochondrion. 25, 17–27 (2015).
Smith, P. M. et al. The role of the mitochondrial ribosome in human disease: searching for mutations in 12S mitochondrial rRNA with high disruptive potential. Hum. Mol. Genet. 23, 949–967 (2014).
Wallace, D. C., Lott, M. T., & Procaccio, V. Mitochondrial medicine and biology. Emery and Rimoin’s principle and practice of medical genetics and genomics (7th edn) 267–322 (Elsevier Science, 2018).
Lehmann, D. et al. Understanding mitochondrial DNA maintenance disorders at the single muscle fibre level. Nucleic Acids Res. 47, 7430–7443 (2019).
Rossignol, R. et al. Mitochondrial threshold effects. Biochem J. 370, 751–762 (2003).
Wong, L. J. Next generation molecular diagnosis of mitochondrial disorders. Mitochondrion. 13, 379–387 (2013).
Fayssoil, A. et al. Prediction of long-term prognosis by heteroplasmy levels of the m.3243A>G mutation in patients with the mitochondrial encephalomyopathy, lactic acidosis and stroke-like episodes syndrome. Eur. J. Neurol. 24, 255–261 (2017).
Liu, H. et al. Wild-type mitochondrial DNA copy number in urinary cells as a useful marker for diagnosing severity of the mitochondrial diseases. PLoS One. 8, e67146 (2013).
Dong, D. W. et al. Association of G-quadruplex forming sequences with human mtDNA deletion breakpoints. BMC Genomics. 15, 677 (2014).
El-Hattab, A. W., Craigen, W. J. & Scaglia, F. Mitochondrial DNA maintenance defects. Biochim. Biophys. Acta Mol. Basis Dis. 1863, 1539–1555 (2017).
Damas, J. et al. Mitochondrial DNA deletions are associated with non-B DNA conformations. Nucleic Acids Res. 40, 7606–7621 (2012).
Sadikovic, B. et al. Sequence homology at the breakpoint and clinical phenotype of mitochondrial DNA deletion syndromes. PLoS One. 5, e15687 (2010).
Chan, S. S. & Copeland, W. C. DNA polymerase gamma and mitochondrial disease: understanding the consequence of POLG mutations. Biochim. Biophys. Acta. 1787, 312–319 (2009).
Rahman, S. & Copeland, W. C. POLG-related disorders and their neurological manifestations. Nat. Rev. Neurol. 15, 40–52 (2019).
Goudenege, D. et al. eKLIPse: a sensitive tool for the detection and quantification of mitochondrial DNA deletions from next-generation sequencing data. Genet. Med. 21, 1407–1416 (2019).
Richards, S. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405–424 (2015).
Boucret, L. et al. Deep sequencing shows that oocytes are not prone to accumulate mtDNA heteroplasmic mutations during ovarian ageing. Hum. Reprod. 32, 2101–2109 (2017).
Young, M. J. & Copeland, W. C. Human mitochondrial DNA replication machinery and disease. Curr. Opin. Genet. Dev. 38, 52–62 (2016).
Damas, J. et al. rearrangements in health and disease-a comprehensive study. Hum. Mutat. 35, 1–14 (2014).
Diot, A., Morten, K. & Poulton, J. Mitophagy plays a central role in mitochondrial ageing. Mamm. Genome. 27, 381–395 (2016).
Wong, L. J. Recognition of mitochondrial DNA deletion syndrome with non-neuromuscular multisystemic manifestation. Genet. Med. 3, 399–404 (2001).
Grady, J. P. et al. Disease progression in patients with single, large-scale mitochondrial DNA deletions. Brain. 137, 323–334 (2014).
Raymond, F. L., Horvath, R. & Chinnery, P. F. firstline genomic diagnosis of mitochondrial disorders. Nat. Rev. Genet. 19, 399–400 (2018).
Rodriguez-Lopez, C. et al. Clinical, pathological and genetic spectrum in 89 cases of mitochondrial progressive external ophthalmoplegia. J. Med. Genet. 57, 643–646 (2020).
Dorrenhaus, A. et al. Cultures of exfoliated epithelial cells from different locations of the human urinary tract and the renal tubular system. Arch. Toxicolo. 74, 618–626 (2000).
Grady, J. P. et al. mtDNA heteroplasmy level and copy number indicate disease burden in m.3243A>G mitochondrial disease. EMBO Mol. Med. 10, e8262 (2018).
Nissanka, N., Minczuk, M. & Moraes, C. T. Mechanisms of mitochondrial DNA deletion formation. Trends Genet. 35, 235–244 (2019).
Varhaug, K. N. et al. Using urine to diagnose large-scale mtDNA deletions in adult patients. Ann. Clin. Transl. Neurol. 7, 1318–1326 (2020).
Lauber, J., Marsac, C., Kadenbach, B. & Seibel, P. Mutations in mitochondrial tRNA genes: a frequent cause of neuromuscular diseases. Nucleic Acids Res. 19, 1393–1397 (1991).
Moraes, C. T. et al. Mitochondrial DNA deletions in progressive external ophthalmoplegia and Kearns–Sayre syndrome. N. Engl. J. Med. 320, 1293–1299 (1989).
Rocha, M. C. et al. Pathological mechanisms underlying single large-scale mitochondrial DNA deletions. Ann. Neurol. 83, 115–130 (2018).
López-Gallardo, E., López-Pérez, M. J., Montoya, J. & Ruiz-Pesini, E. CPEO and KSS differ in the percentage and location of the mtDNA deletion. Mitochondrion. 9, 314–317 (2009).
Grady, J. P. et al. Accurate measurement of mitochondrial DNA deletion level and copy number differences in human skeletal muscle. PLoS One. 9, e114462 (2014).
Reeve, A. K. et al. Nature of mitochondrial DNA deletions in substantia nigra neurons. Am. J. Hum. Genet. 82, 228–235 (2008).
Schon, K. R., Ratnaike, T., van den Ameele, J., Horvath, R. & Chinnery, P. F. Mitochondrial diseases: a diagnostic revolution. Trends Genet. 36, 702–717 (2020).
Acknowledgements
This work was supported by grants from Association contre les Maladies Mitochondriales (AMMi), AFM-Telethon, Aviesan INSERM Genomic Variability Project, and from the Angers University Hospital. We thank all members of the French mitochondrial disease network (MITODIAG) for their support.
Author information
Authors and Affiliations
Contributions
Conceptualization: C.B., D.G., V.P. Formal analysis and data curation: V.D.D., N.G., M.A., F.L., G.L, P.R., P.A.B., C.B., D.G., V.P. Investigation and resources: S.B., P.G., B.R., A.T., S.A., C.R., S.S., C.J., A.S.., M.B., C.V., M.S., J.C., E.C., Y.P., M.L.M.N., V.P.F., D.B. Writing—original draft V.P., C.B., G.L. Writing—review & editing: input from all authors, who approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics declaration
DNA samples from controls and patients with mitochondrial disease, analyzed both retrospectively and prospectively, were collected after obtaining written informed consent (IRB, University Hospital of Angers, DC2014-2224).
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Bris, C., Goudenège, D., Desquiret-Dumas, V. et al. Improved detection of mitochondrial DNA instability in mitochondrial genome maintenance disorders. Genet Med 23, 1769–1778 (2021). https://doi.org/10.1038/s41436-021-01206-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-021-01206-w
This article is cited by
-
OxPhos defects cause hypermetabolism and reduce lifespan in cells and in patients with mitochondrial diseases
Communications Biology (2023)