Abstract
Purpose
Variant classifications and gene–disease relationships may evolve. Professional societies have suggested patients share the responsibility to remain up-to-date on the implications genetic results have on their health, and that novel methods of recontact are needed. GenomeConnect, the ClinGen patient registry, has implemented a process to provide variant classification and gene–disease relationship updates to participants. Here, we report on our experience with this recontacting process.
Methods
GenomeConnect shares data with ClinVar and Matchmaker Exchange enabling the identification of updates to variant classifications and gene–disease relationships. For any updates identified, the reporting laboratory is contacted, and updates are shared with participants opting to receive them.
Results
Of 1,419 variants shared with ClinVar by GenomeConnect, 49 (3.4%) variant reclassifications were identified and 34 were shared with participants. Of 97 candidate genes submitted to Matchmaker Exchange, 10 (10.3%) gene–disease relationships have been confirmed and 9 were shared with participants. Details available from a subset of participants highlight that updated information is not always shared with the patient by testing laboratories.
Conclusion
Patient registries can provide a mechanism for patients and their providers to remain informed about changes to the interpretation and clinical significance of their genetic results, leading to important implications for care.
Graphical Abstract

Similar content being viewed by others
INTRODUCTION
Genetic advances, including increased availability of next-generation sequencing, improved variant classification guidelines, enhanced population databases, and expanded data sharing efforts, are enabling genomic discovery. As discoveries are made and evidence is accumulated, variant classifications and gene–disease relationships can shift, resulting in potential updates to testing results, some of which will impact medical management.1,2,3
Clinical laboratories, researchers, and clinicians’ duty to reassess genomic results and recontact patients has been the subject of ongoing discussion. Although no legal duty to recontact has been established, there is consensus that recontact is beneficial and that most patients value receiving updated information.4,5 The American College of Medical Genetics and Genomics (ACMG) suggests that laboratories do not have a duty to reanalyze data unless requested, but should revisit variant classifications in several instances, including when new evidence (e.g., genomic resource and publications supporting new gene–disease relationships) or updated variant assessment methods become available.6,7 ACMG acknowledges that when a variant is reclassified, it is “good clinical practice” for laboratories to identify patients with that variant in their database and issue updated reports to referring providers.7 They also recognize that laboratories have limited resources to support this practice without reimbursement.8 The European Society of Human Genetics (ESHG) also proposes that laboratories are not required to reanalyze sequencing data, but acknowledge a responsibility for laboratories to reissue reports and recontact referring clinicians should a variant be reclassified.9 Reassessment and recontact protocols at testing laboratories vary.10 Moreover, the feasibility of continued variant reassessment by the reporting laboratory has been questioned.6,8,11
Clinicians’ recontacting practices also vary with few standards to guide practice.12,13 While genetics providers see recontact as desirable,5 and a majority of clinics appear to recontact to some degree, few do so routinely.12,13 Clinicians that receive updated information from the testing laboratory or identify novel information in their own reassessment can encounter barriers to recontacting patients including lack of time, resources, and current patient contact information.5 Clinical reimbursement for variant reassessment and improved infrastructure to enable recontact are needed to ensure patients can remain informed about their genetic test results.4,6 Under current circumstances, however, engaging the patient in the responsibility of recontact has been suggested as a means to ensure that patients are receiving up-to-date information.4,5,7,14
In the research setting, the American Society of Human Genetics (ASHG) “strongly recommends” recontact if research included return of results and a reinterpretation of genomic information results in an update that is related to the phenotype being studied or would impact management.15 ASHG indicates that recontact may be considered in other situations, including in cases where the update is unlikely to impact care, but asserts that researchers are not expected to scan genetic data or literature for updated information, and the responsibility to recontact is limited to the duration of funding.15
GenomeConnect, the NIH-funded Clinical Genome Resource (ClinGen)16 patient registry, was launched in October 2014 as a mechanism for patients who have had genetic testing to contribute their genetic and health data to publicly available databases to inform the community’s understanding of genetics and health.17,18 Recognizing that the registry could identify updates to participants’ genetic test results through its data sharing practices, GenomeConnect implemented a process to provide genetic updates back to interested participants in February 2017.18 Here, we report on our experience over the past three and a half years providing variant classification and gene–disease validity updates to registry participants.
MATERIALS AND METHODS
Participants
GenomeConnect is approved by the Geisinger Institutional Review Board (2014-0408). Informed consent for data sharing and recontact is obtained from all participants or participants’ parent/legal guardians, and electronic assent is obtained from participants ages 10–17.
Data sharing
As described previously, participant-provided phenotype information and classified genomic variants collected by registry staff from participants’ genetic test reports are de-identified and shared with ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/submitters/506185/).19 Variants in genes without a clear relationship to a Mendelian disease (hereafter referred to as “candidate genes”) are submitted to Matchmaker Exchange (MME)20 through GeneMatcher.17,18,21 Prior to submitting a gene to GeneMatcher, searches of OMIM and ClinGen Gene–Disease Validity curations22 (https://search.clinicalgenome.org/kb/gene-validity/) are completed to explore if a gene–disease relationship has been identified since the variant was initially reported.
Recontacting process
In February 2017, GenomeConnect established a process to return updates to participants.18 Upon enrollment, individuals are required to provide their preference regarding receipt of updates from the registry. Participants that enrolled prior to February 2017 were prompted to update their preferences via email and reminders were included in the registry’s quarterly newsletters.
Using ClinVar’s weekly data release, ClinGen generates reports comparing variants submitted by GenomeConnect to other ClinVar submitters’ data. These reports enable tracking of processed variants and are used to identify discrepancies between GenomeConnect submissions and the reporting laboratory’s classification, if available, to enable ongoing assessment for potential updates. Potential updates (i.e., when the reporting laboratory’s assessment of the variant in ClinVar differs from the assessment on the participant’s report) are identified following a process previously outlined.18
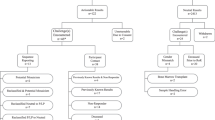
Additionally, the registry might identify potential updates to gene–disease validity through its submission to MME. Potential updates can be identified prior to submission when the registry team searches OMIM and ClinGen for novel gene–disease relationships or following submission if a subsequent publication identifies a possible novel disease relationship (Fig. 1). Per ACMG/Association for Molecular Pathology (AMP) sequence variant interpretation guidelines, an established gene–disease relationship is necessary to determine a variant’s pathogenicity, and, without such, a variant should be classified as a variant of uncertain significance (VUS).8 Establishing a gene–disease relationship can enable a variant to be reassessed and potentially upgraded to likely pathogenic/pathogenic. Unlike variant classification updates identified through ClinVar, gene–disease relationship updates identified through MME do not provide information regarding the reporting laboratory’s current classification. In instances where a potential disease relationship is identified, the registry team reaches out to the reporting laboratory to inquire if such information would result in a variant reclassification (Fig. 1). Participants are then recontacted on a case-by-case basis depending on the laboratory’s response (e.g., if the laboratory now considers the gene to have a Mendelian association, the registry would contact the participant).
After variant classification or gene–disease updates are identified, participants are contacted, as previously described, via the email address associated with their GenomeConnect account and informed there might be updated information regarding their genetic test results.18 The specific gene that the update pertains to is included in the email, and, in the case of variant reclassifications, the notification includes a link to the ClinVar record (Supplementary Materials and Methods). Discussion of the nature of the update and impact on care, if any, are best conducted within the provider–patient relationship; because the registry is unable to assess clinical correlation or recommend appropriate management, the notification does not provide a complete summary of the updated information.18 Participants are directed to contact the provider that ordered their testing or a genetics professional in their area for discussion regarding potential updates. Follow-up information after an update is not systematically obtained for all participants but is documented, if provided.
Descriptive statistics are summarized as median and range or frequency and percentage as appropriate.
RESULTS
From October 2014 to October 2020, 3,134 participants enrolled in GenomeConnect. Prior to February 2017, participants were not asked about their preference regarding updates when enrolling in the registry. Since adding a question about such preferences, 97.7% (n = 2,154/2,205) of participants have opted to receive updates. Of the 929 participants that enrolled prior to February 2017, 80 (8.6%) have provided their preference regarding receipt of updates; of those, 93.8% (n = 75) opted to receive updates. Of those that opted in to receiving updates, 38.6% (n = 860/2,229) shared a genetic testing report that could be reviewed and, if any variants were identified, shared with external databases.
ClinVar submission
As of October 2020, 1,419 variants from 674 GenomeConnect participants are publicly available within ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/submitters/506185/). Of those 1,419 variants, 585 (41.2%) were novel to the database. The remaining 834 variants have submissions by at least one other institution. Of those, 481 (57.7%) were submitted by the GenomeConnect participant’s reporting laboratory, enabling comparison between the classification on the participant’s report and the laboratory’s most recent ClinVar submission. Forty-nine variant classifications (10.2%, n = 49/481) collected from 47 GenomeConnect participants’ reports were out-of-date compared to the laboratory’s most recent submission. Two individuals had multiple variant reclassifications identified (one participant had two and a second participant had three). One variant was reported in two related registry participants. Demographic data of the 47 participants with an identified variant classification update compared to that of all participants whose variants have been submitted to ClinVar are summarized in Table 1. These participants’ 47 reports came from ten different clinical laboratories and were issued between May 2011 and March 2018 (median December 2015; Table 1). According to evaluation dates in ClinVar, the laboratories updated their classification a median of 1.63 years after the participants’ initial report (range 0–4.93 years).
Forty-two (85.7%, n = 42/49) discrepancies between the GenomeConnect participant’s report and the reporting laboratory’s ClinVar submission were differences between the major classification categories (pathogenic/likely pathogenic, VUS, and benign/likely benign). Four variants (8.2%) were upgraded from VUS to pathogenic/likely pathogenic. The remaining 38 major category reclassifications were downgrades. Thirty-four variants were downgraded from VUS to benign/likely benign (69.4%), two from pathogenic to VUS (4.1%), and two from pathogenic to benign/likely benign (4.1%). Only seven differences (14.3%) were due to changes in the level of confidence (e.g., likely benign versus benign or likely pathogenic versus pathogenic) (Table S1).
Of the 49 variant classification updates, 34 (69.4%) were shared back with participants who opted to receive them (Fig. 2). The remaining 15 updates (30.6%) have not been provided to participants because they have not made a selection regarding receipt of updates.
MME submission
Of the 97 genes reported as candidate genes on participant reports (n = 65 participants), 4 genes were found to have a potential Mendelian relationship prior to MME submission through a brief search of OMIM and ClinGen curations. The remaining 93 genes were submitted to MME. After submission, a match identified through MME provided a recent publication documenting a newly described disease relationship for seven additional genes. Articles shared by an MME match were published an average of 2.08 years (range -0.25–3.93 years) after the participants’ testing. For two additional genes, GenomeConnect discovered literature suggesting a disease relationship following MME submission. None of the variants in these 13 genes were submitted to ClinVar by the reporting laboratory at the time potential gene–disease relationship updates were identified.
For all 13 genes with potential novel gene–disease relationships, the reporting laboratory was contacted to inquire about their current gene–disease validity classification (Fig. 3). Three genes (ACKR3, NSF, TAOK1) are still considered candidate genes by the reporting laboratory. The remaining ten genes are now considered to be associated with a Mendelian disease, resulting in updates for ten participants. Demographic data of participants with an identified gene–disease relationship update as compared to all participants who had a candidate gene submitted to MME are summarized in Table 2.
The participants’ initial reports were released between July 2012 and June 2019 (median August 2017). From discussion with the reporting laboratory, GenomeConnect learned that reports for three participants had been updated because of the new gene–disease relationship prior to GenomeConnect’s contact (HECW2, MSL3, SON; Fig. 3). The gene–disease relationship for a fourth participant (ASXL3) had been updated prior to the registry’s contact with the reporting laboratory, but an updated report has not been issued to date. For the six remaining participants, gene–disease relationships and reports were updated by the laboratory after GenomeConnect’s contact (BCL11B, CSNK2B, GRIA2, KDM3B, ZNF292, TRPM3). Of the ten potential gene–disease relationship updates, 9 (90.0%) were shared back with participants that opted to receive them. The participant with the ASXL3 variant enrolled prior to February 2017 and has not made a selection regarding receipt of updates.
All variants in these ten genes were initially reported as VUS. As a result of these gene-level updates, the pathogenicity of the ten variants could be reassessed by the reporting laboratory. Variants in the nine genes with gene–disease relationship updates returned to participants (BCL11B, CSNK2B, GRIA2, KDM3B, ZNF292, TRPM3, MSL3, HECW2, and SON) were reclassified to pathogenic/likely pathogenic by the reporting laboratories. The variant in ASXL3 has not been reassessed by the reporting laboratory to date since they require a request from the ordering provider to do so (Fig. 3).
Case examples
Although follow-up information is not available for all participants who received an update, the following selected case examples illustrate the effect of identifying and sharing variant reclassifications and gene–disease relationship updates through a patient registry.
Case 1: Variant Reclassified After Identification in Additional Patient
Case 1 underwent exome sequencing (ES) in 2016 due to a history of seizures and cognitive impairment. A variant in CSNK2B, a neurodevelopmental candidate gene, was identified. After submission to MME, additional evidence suggesting a potential relationship between CSNK2B, intellectual disability, and seizures was described in the literature.23 GenomeConnect contacted the reporting laboratory and learned that, since the original report, the gene had been upgraded to disease-causing, and the laboratory had observed the variant in a subsequent patient and reported it as pathogenic. Since the initial report, case 1’s family had been in contact with the reporting laboratory to request a stored DNA sample for research purposes. However, the variant reclassification had not been shared with this patient or their clinician prior to GenomeConnect’s inquiry. GenomeConnect’s contact with the reporting laboratory prompted them to update the patient’s report and notify the ordering provider of the reclassification. An updated report coupled with GenomeConnect’s contact enabled the family to receive this new information.
Case 2: Updated Classification Shared with Clinician Previously
In 2015, case 2 underwent ES due to a history of global developmental delay, encephalopathy, seizures, hypotonia, failure to thrive, and cardiac anomalies. A de novo missense variant in HECW2, a neurodevelopmental candidate gene, was reported. Prior to MME submission, a disease relationship was noted in OMIM (OMIM 617268; page created 21 December 2016). The registry contacted the reporting laboratory and was informed that the variant had been upgraded to pathogenic based on the novel gene–disease relationship, and an updated report had been sent to the referring clinician in March 2018. In keeping with standard procedures for returning updates, GenomeConnect contacted the family with the update in December 2019. At that time, the family shared they had not received an updated report and planned to contact their clinician to get access to the report and discuss its implications.
Case 3: Variant Reclassification Prompting Additional Reassessment
Case 3 underwent ES in 2015 due to a history of prenatal cystic hygroma, behavioral issues, fine motor delay, and dysmorphisms. Several variants were identified including biallelic variants in DHCR7, the gene associated with Smith–Lemli–Opitz syndrome (OMIM 270400). One was interpreted as pathogenic and the second as a VUS. Through ClinVar, GenomeConnect identified that the VUS had been downgraded by the reporting laboratory to likely benign. After being contacted by GenomeConnect with this update, the patient underwent ES reanalysis and received a report documenting this reclassification and identification of a novel, de novo VUS in another gene associated with dysmorphic features, neurodevelopmental disorders, and congenital anomalies. The update shared by GenomeConnect prompted this reassessment and further investigation into the genetic etiology for the participant’s symptoms.
Case 4: Case Series Established a gGene–disease relationship
Case 4 underwent ES in 2016 due to a history of neurodevelopmental phenotypes including autism spectrum disorder, developmental delay, regression, Tourette syndrome, and hypotonia. VUS in two candidate genes, GRIA1 and GRIA2, were identified. GenomeConnect submitted these candidate genes to MME, and, subsequently, a match led to identification of a recent publication suggesting a relationship between GRIA2 and neurodevelopmental phenotypes.24 GenomeConnect reached out to the reporting laboratory and learned that, given this new information, the gene would now be considered to have a valid disease relationship and the variant would be reclassified as likely pathogenic. The gene and variant had not been reclassified prior to the registry’s inquiry.
Case 5: participant enrolled to remain informed about a VUS
Case 5 underwent panel testing following an aortic dissection in 2015. A VUS in FLNA, a gene associated with a spectrum of X-linked phenotypes (OMIM 300017), was identified. The participant enrolled in GenomeConnect to “stay abreast of any developments regarding this varian[t].” In 2018, the reporting laboratory reclassified the variant as likely benign and updated their ClinVar submission. The participant was contacted, informed that there was a potential update to their results, and directed to their provider for further discussion. Although the participant is still without a genetic explanation for their clinical history, by enrolling in GenomeConnect, they remained informed about their results.
DISCUSSION
Advances in genomic medicine will identify variant classification and gene validity updates. ACMG and ESHG have highlighted that recontact is a shared responsibility between patients, clinicians, and laboratories.4,5,7,14 Additionally, involvement of other stakeholders, including patient associations, has been suggested to ensure patients remain informed.4 As of October 2020, GenomeConnect has provided 43 total updates back to interested participants—34 variant reclassifications and 9 gene–disease relationship updates. These updates were provided to 40 participants in total (representing 4.7% of participants who uploaded a genetic test report and opted to receive updates), illustrating that patient registries can provide an innovative way for patients to remain informed.
Although outcomes are not available for all participants who received an update, from those with follow-up information, it is apparent that patients are not always receiving updates in current practice. In cases where the laboratory and/or clinician are aware of updated information, patients might not be informed due to variable recontacting practices and barriers to recontact. GenomeConnect participants were unaware of updates to their results even when their variant had been reinterpreted by their reporting laboratory (case 1) or their clinician had received an updated report (case 2). As illustrated by the six gene–disease relationship reassessments prompted by GenomeConnect, the reporting laboratory or clinician might be unaware of novel literature or resources that could prompt updated classifications of variants and gene–disease relationships.
Based on GenomeConnect’s experience to date, we anticipate that the registry will continue to identify updates moving forward. The majority of variants that were reclassified were initially reported after the publication of the ACMG/AMP sequence variant interpretation guidelines (n = 41/49, 83.7%)8 and after the launch of the Exome Aggregation Consortium (ExAC) browser (n = 43/49, 87.8%).24 Over one-third were reported after the launch of the Genome Aggregation Database (gnomAD) (n = 17/49, 34.7%).25 These data suggest that the availability of these resources alone does not account for the variant reclassifications identified by GenomeConnect. Additionally, these resources would not be expected to influence upgrades from VUS to P/LP, which largely come from new cases and published studies. Furthermore, increasing availability and use of exome and genome sequencing will result in a growing number of variants in candidate genes being reported to patients and subsequent gene–disease discoveries in the coming years.26
Consistent with previous reports from data in ClinVar and clinical laboratories, the largest proportion of variant reclassifications identified by GenomeConnect were downgrades (77.6%, n = 38/49), and, in particular, downgrades from VUS to benign/likely benign (n = 34/49).2,3,27,28,29 These, along with confidence reclassifications (e.g., likely pathogenic to pathogenic), are unlikely to have a significant impact on patient care, but can reduce uncertainty and, in some cases, stimulate further investigation of other causes of disease. Given current barriers to recontact, laboratories and clinicians prioritize reassessment and recontact based on potential clinical and/or personal utility.4,6,12,15 At least a subset of patients, however, want any type of novel information.18,30 Based on GenomeConnect’s experience returning updates to participants, even updates that might not have clear clinical implications, such as the VUS to likely benign update in case 3, can prompt additional assessments that could inform diagnoses. By using novel methods, such as patient registries for recontact, patients can be informed of updates that might not be prioritized by laboratories and/or clinicians under current constraints.
A limitation of this study is that detailed follow-up is not available for all participants receiving an update. Future studies should attempt to capture follow-up details for all participants to better understand the impact of patient registries providing updates to participants. Given previous reports of limited patient follow-up,4,31 variable understanding,4,32,33 and diverse psychosocial reactions28,32,33,34 after recontact, additional studies are needed to capture psychosocial outcomes, barriers to follow-up with a clinician, changes to diagnoses and care, and participant perspectives following recontact by a registry. Such studies, including those employing a randomized design, are needed fully assess recontact by patient registries compared to that by laboratories, researchers, and clinicians.
GenomeConnect’s ability to provide variant classification and gene–disease relationship updates relies on data sharing. If laboratories do not share variant classifications with ClinVar and stakeholders do not collaborate to assess candidate genes, GenomeConnect will not be aware of changes to participants’ variant classifications and updates to gene–disease relationships. Continued efforts by professional societies,35,36,37 medical institutions,38 and payers39 to encourage data sharing will be important. Not only can data sharing inform the genetics community’s understanding of genes and variants, but, as this study shows, it can also enable patients to remain informed about their genetic results.
Moving forward, GenomeConnect will also iterate on the process of providing updates and will explore additional updates that can be provided through the registry. Given our experience receiving recent publications documenting possible disease relationships from matches for seven genes submitted to MME to date, GenomeConnect will begin completing a literature search prior to submission in addition to searches of OMIM and ClinGen curations.
While we have demonstrated that patient registries can play a role in ensuring that patients remain informed about their genetic results, they cannot serve as the only solution to this complex problem. Because of the inherent challenges associated with attempting to recontact individuals, multiple approaches involving laboratories, researchers, clinicians, and registries will be needed to disseminate updated information. GenomeConnect is working to scale engagement of patients in data sharing and provision of genetic testing updates by involving other patient registries in this process. The protocols, data collection, and sharing tools GenomeConnect employs are available to other organizations interested in engaging patients in data sharing and enabling genetic testing updates. Since 2018, GenomeConnect has partnered with ten external patient registries,19 and as of October 2020, 1,126 individuals have chosen to participate in data sharing and were given the option to receive updates through these registries.
Variant classifications and gene–disease relationships reported to patients are based on the information available at the time of report. As advances in genomic medicine are made, updates to results are identified. Although recontacting patients with updated information is considered beneficial, barriers exist preventing updates from reaching patients. GenomeConnect’s experience highlights that patient registries can supplement laboratory and clinicians’ efforts to keep patients informed about their genetic results and stresses the importance of data sharing in identifying updates and making discoveries.
Data availability
GenomeConnect submits variants from participant genetic test reports to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/submitters/506185/).
References
Marcus, A. D. A genetic test led seven women in one family to have major surgery. Then the odds changed. The Wall Street Journal. https://www.wsj.com/articles/seven-women-in-a-family-chose-surgery-after-a-genetic-test-then-the-results-changed-11576860210 (2019).
Macklin, S., Durand, N., Atwal, P. & Hines, S. Observed frequency and challenges of variant reclassification in a hereditary cancer clinic. Genet. Med. 20, 346–350 (2018).
Mersch, J. et al. Prevalence of variant reclassification following hereditary cancer genetic testing. JAMA. 320, 1266–1274 (2018).
Carrieri, D. et al. Recontacting patients in clinical genetics services: recommendations of the European Society of Human Genetics. Eur. J. Hum. Genet. 27, 169–182 (2019).
Otten, E. et al. Is there a duty to recontact in light of new genetic technologies? A systematic review of the literature. Genet. Med. 17, 668–678 (2015).
Deignan, J. L. et al. Points to consider in the reevaluation and reanalysis of genomic test results: a statement of the American College of Medical Genetics and Genomics (ACMG). Genet. Med. 21, 1267–1270 (2019).
Vears, D. F. et al. Points to consider for laboratories reporting results from diagnostic genomic sequencing. Eur. J. Hum. Genet. 26, 36–43 (2018).
Richards, S. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405–424 (2015).
Matthijs, G. et al. Guidelines for diagnostic next-generation sequencing. Eur. J. Hum. Genet. 24, 2–5 (2016).
Vears, D. F., Niemiec, E., Howard, H. C. & Borry, P. Analysis of VUS reporting, variant reinterpretation and recontact policies in clinical genomic sequencing consent forms. Eur. J. Hum. Genet. 26, 1743–1751 (2018).
Aronson, S. J. et al. Communicating new knowledge on previously reported genetic variants. Genet. Med. 14, 713–719 (2012).
Carrieri, D. et al. Recontact in clinical practice: a survey of clinical genetics services in the United Kingdom. Genet. Med. 18, 876–881 (2016).
Sirchia, F. et al. Recontacting or not recontacting? A survey of current practices in clinical genetics centres in Europe. Eur. J. Hum. Genet. 26, 946–954 (2018).
David, K. L. et al. Patient re-contact after revision of genomic test results: points to consider—a statement of the American College of Medical Genetics and Genomics (ACMG). Genet. Med. 21, 769–771 (2019).
Bombard, Y. et al. The responsibility to recontact research participants after reinterpretation of genetic and genomic research results. Am. J. Hum. Genet. 104, 578–595 (2019).
Rehm, H. L. et al. ClinGen—the Clinical Genome Resource. N. Engl. J. Med. 372, 2235–2242 (2015).
Kirkpatrick, B. E. et al. GenomeConnect: matchmaking between patients, clinical laboratories, and researchers to improve genomic knowledge. Hum. Mutat. 36, 974–978 (2015).
Savatt, J. M. et al. ClinGen’s GenomeConnect registry enables patient-centered data sharing. Hum. Mutat. 39, 1668–1676 (2018).
Landrum, M. J. et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 46, D1062–D1067 (2018).
Philippakis, A. A. et al. The Matchmaker Exchange: a platform for rare disease gene discovery. Hum. Mutat. 36, 915–921 (2015).
Sobreira, N., Schiettecatte, F., Valle, D. & Hamosh, A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 36, 928–930 (2015).
Strande, N. T. et al. Evaluating the clinical validity of gene–disease associations: an evidence-based framework developed by the Clinical Genome Resource. Am. J. Hum. Genet. 100, 895–906 (2017).
Nakashima, M. et al. Identification of de novo CSNK2A1 and CSNK2B variants in cases of global developmental delay with seizures. J. Hum. Genet. 64, 313–322 (2019).
Salpietro, V. et al. AMPA receptor GluA2 subunit defects are a cause of neurodevelopmental disorders. Nat. Commun. 10, 3094 (2019).
Anderson, A. GnomAD resource introduced at ASHG meeting, doubles ExAC dataset. GenomeWeb. https://www.genomeweb.com/sequencing/gnomad-resource-introduced-ashg-meeting-doubles-exac-dataset#.Xs6nf4hKgdU (2016).
Bamshad, M. J., Nickerson, D. A. & Chong, J. X. Mendelian gene discovery: fast and furious with no end in sight. Am. J. Hum. Genet. 105, 448–455 (2019).
Harrison, S. M. & Rehm, H. L. Is ‘likely pathogenic’ really 90% likely? Reclassification data in ClinVar. Genome Med. 11, 72 (2019).
Slavin, T. P. et al. Prospective study of cancer genetic variants: variation in rate of reclassification by ancestry. J. Natl. Cancer Inst. 110, 1059–1066 (2018).
Schultz, C., Blanco, K., Powis, Z. & Ichikawa, S. Has anything changed? Reclassification of epilepsy genetic variants. Poster presented at the 2019 ACMG Clinical Genetics Meeting, Seattle, 4 April 4 2019. Poster 715.
Carrieri, D. et al. Recontacting in clinical practice: the views and expectations of patients in the United Kingdom. Eur. J. Hum. Genet. 25, 1106–1112 (2017).
Romero Arenas, M. A. et al. Recontacting patients with updated genetic testing recommendations for medullary thyroid carcinoma and pheochromocytoma or paraganglioma. Ann. Surg. Oncol. 25, 1395–1402 (2018).
Halverson, C. M. E., Connors, L. M., Wessinger, B. C., Clayton, E. W. & Wiesner, G. L. Patient perspectives on variant reclassification after cancer susceptibility testing. Mol. Genet. Genomic Med. 8, e1275 (2020).
Wong, E. K. et al. Perceptions of genetic variant reclassification in patients with inherited cardiac disease. Eur. J. Hum. Genet. 27, 1134–1142 (2019).
Beunders, G., Dekker, M., Haver, O., Meijers-Heijboer, H. J. & Henneman, L. Recontacting in light of new genetic diagnostic techniques for patients with intellectual disability: feasibility and parental perspectives. Eur. J. Med. Genet. 61, 213–218 (2018).
American Medical Association. Genome analysis and variant identification Policy D-460.971. https://policysearch.ama-assn.org/policyfinder/search/Genome%20Analysis%20and%20Variant%20Identification%20D-460.971/relevant/1/ (2013).
National Society of Genetic Counselors (NSGC). Clinical data sharing, 2020. https://www.nsgc.org/p/bl/et/blogaid=330 (2020).
American College of Medical Genetics and Genomics Board of Directors. Laboratory and clinical genomic data sharing is crucial to improving genetic health care: a position statement of the American College of Medical Genetics and Genomics. Genet. Med. 19, 721–722 (2017).
Geisinger. Genomic Medicine Institute. https://www.geisinger.edu/research/departments-and-centers/gmi (2017).
Ray, T. Genomic variant data sharing gains support; collaboration seen as key to interpretation challenge. GenomeWeb. https://www.genomeweb.com/informatics/genomic-variant-data-sharing-gains-support-collaboration-seen-key-interpretation#.Xs_eGjZYYdU (2016).
Acknowledgements
This work was supported by the National Human Genome Research Institute of the National Institutes of Health (NIH) under award number U41HG006834. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The authors acknowledge the GenomeConnect participants as well as the laboratories that reported variants to these participants and worked with GenomeConnect to update reports. GenomeConnect is built on the Invitae Patient Insights Network platform. The authors also acknowledge the Invitae team for their work in building and maintaining GenomeConnect.
Author contributions
H.L.R., C.L.M., D.H.L.: Funding acquisition; J.M.S., D.R.A., D.H.L., H.L.R., E.R.R., C.L.M.: Conceptualization; J.M.S., D.R.A., H.L.R., E.R.R., C.L.M.: Methodology; E.R.R., C.L.M.: Project administration; E.R.R., C.L.M.: Supervision; J.M.S., E.P.: Data curation; J.M.S.: Investigation, Formal analysis, Visualization; J.M.S., E.R.R., D.R.A., C.L.M.: Writing—original draft; J.M.S., D.R.A., D.H.L., E.P., H.L.R., E.R.R., C.L.M.: Writing—review & editing.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics Declaration
GenomeConnect is approved by the Geisinger Institutional Review Board (2014-0408). Consent is obtained from participants or participants’ parent/legal guardian, and electronic assent is obtained from participants ages 10–17. Data are de-identified prior to submission to ClinVar and other databases. Individual data included in this paper have been de-identified.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Savatt, J.M., Azzariti, D.R., Ledbetter, D.H. et al. Recontacting registry participants with genetic updates through GenomeConnect, the ClinGen patient registry. Genet Med 23, 1738–1745 (2021). https://doi.org/10.1038/s41436-021-01197-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-021-01197-8
This article is cited by
-
The Brain Gene Registry: a data snapshot
Journal of Neurodevelopmental Disorders (2024)
-
Exploring the impact of the reclassification of a hereditary cancer syndrome gene variant: emerging themes from a qualitative study
Journal of Community Genetics (2023)
-
“There should be one spot that you can go:” BRCA mutation carriers’ perspectives on cancer risk management and a hereditary cancer registry
Journal of Community Genetics (2023)
-
Cancer patients’ understandings of genetic variants of uncertain significance in clinical care
Journal of Community Genetics (2022)
-
Fragmented responsibility: views of Israeli HCPs regarding patient recontact following variant reclassification
Journal of Community Genetics (2022)