Abstract
Purpose
Secondary findings are typically offered in an all or none fashion when sequencing is used for clinical purposes. This study aims to describe the process of offering categorical and granular choices for results in a large research consortium.
Methods
Within the third phase of the electronic MEdical Records and GEnomics (eMERGE) Network, several sites implemented studies that allowed participants to choose the type of results they wanted to receive from a multigene sequencing panel. Sites were surveyed to capture the details of the implementation protocols and results of these choices.
Results
Across the ten eMERGE sites, 4664 participants including adolescents and adults were offered some type of choice. Categories of choices offered and methods for selecting categories varied. Most participants (94.5%) chose to learn all genetic results, while 5.5% chose subsets of results. Several sites allowed participants to change their choices at various time points, and 0.5% of participants made changes.
Conclusion
Offering choices that include learning some results is important and should be a dynamic process to allow for changes in scientific knowledge, participant age group, and individual preference.
Similar content being viewed by others
INTRODUCTION
Genetic and genomic testing has become more common in clinical and research settings. When genes beyond clinical or research indication are purposefully assessed for potential return, analyses are considered opportunistic and the findings secondary. Studies have demonstrated overwhelming support by research participants and the general public for the return of these secondary findings; yet some individuals want only a subset of secondary findings or do not want to receive any.1,2,3,4,5,6,7,8,9,10,11 Providing those undergoing clinical or research genomic testing with options for which secondary findings they would like returned is respectful of patient autonomy but may be logistically challenging.
Special considerations need to be made for children and adolescents undergoing genome testing, since some secondary findings may not manifest until adulthood. Genetic, pediatric, and ethics experts have recommended against testing children for adult-onset conditions to preserve a child’s future autonomy to make their own choices with regard to learning about adult-onset conditions.12,13,14,15 However, studies indicate that the majority of parents want to learn all possible secondary findings results for their children, which may not reflect the current or future perspectives of their children.8,16,17,18 While there has been debate over how involved children and adolescents should be in the consenting/assenting process for the return of secondary findings, there is a growing consensus that, when possible, a collaborative approach is best with continued discussions as the child ages and enters different life stages.19,20
There are a variety of different implementation approaches to returning secondary findings from diagnostic genomic sequencing.21,22,23,24 Clinical genetic testing laboratories offer opt in/opt out or mandatory return of results for all or a subset of genes recommended for opportunistic analysis by the American College of Medical Genetics and Genomics.23,25 Some clinical laboratories provide options for opportunistic analysis of additional genes per individual laboratory policy.25 Different logistical approaches have been suggested for how to categorize genes and associated conditions and how to manage individuals’ choices beyond learning all or none of the potential results. In addition to actionability, examples of genomic result categories that have been suggested or studied include disease severity, age of onset, preventability, treatability, carrier status of autosomal recessive disorders, pharmacogenetic variants, and phenotype.24,25,26,27,28,29,30 Studies have suggested that choices made by individuals undergoing sequencing are influenced by personal experiences and values, and therefore the ability to opt in or out of all possible results is not sufficient.19,30,31,32
While the issue of providing choices for secondary results stems from challenges in the clinical space, it has increasing relevance in the research setting. Clinical standards influence policies for return of genomic results in research.33 However, it is unclear if returning secondary results for a large-scale research project is feasible and warranted. Few studies have tried to prospectively implement choices for discrete categories of genomic results in a large population.34 Here we present different approaches to prospectively provide research participants with options for learning genomic results from panel testing in a multicenter research consortium. This study aims to describe the choices made by participants, as well as the considerations and challenges that shaped the protocols used to implement them.
MATERIALS AND METHODS
Ethics statement: Each site’s protocol, consent process, and consent and assent (if applicable) forms were reviewed and approved by their individual institutional review boards (IRBs). Each site obtained informed consent and assent, when relevant, from their participants.
This study assessed participant choices within the third phase of the eMERGE network funded by the National Human Genome Research Institute. Ten sites, two sequencing centers, and one coordinating center made up the eMERGE III Network. The eMERGE III sites included Cincinnati Children’s Hospital Medical Center (CCHMC), Children’s Hospital of Philadelphia (CHOP), Columbia University (CU), Geisinger (GE), Kaiser Permanente of Washington/University of Washington (KPWA/UW), Mayo Clinic (MC), Meharry Medical College (MMC), Northwestern University (NU), Partners HealthCare (PHC), and Vanderbilt University Medical Center (VUMC). Sites enrolled up to 3000 participants each for a total study population of 25,015.35 Sites independently developed their own recruitment and return of results protocols.36
The sequencing centers, Broad Institute and Partners HealthCare Laboratory for Molecular Medicine, and Baylor College of Medicine Human Genome Sequencing Center, analyzed participant samples using the eMERGEseq panel that contained 109 genes and 1551 single-nucleotide variants (SNVs) chosen by the Network.35 From this panel, the eMERGE Clinical Annotation Workgroup designated 67 genes and 14 SNVs as actionable, and recommended pathogenic and likely pathogenic results be returned to all eMERGE III participants.35 However, each site’s protocol differed regarding which genes and SNVs would be reported.35 A subset of eMERGE sites offered participants a choice to learn all, some, or none of the potential gene results. The term “genomic results” rather than “secondary findings” results is used for the remainder of this paper, as cohorts at some sites were recruited without disease indication.
To understand how sites operationalized choices offered to participants, we developed a questionnaire that was distributed to a designated point of contact at each site within the Network. The questionnaire captured protocol descriptions and data related to participant options. Specifically, sites were asked (1) if they offered their study participants choices, and if so, what categories of choices were offered and how those categories were determined; (2) how genes and SNVs were sorted into these categories; (3) whether or not participants were able to change their selections; and (4) the results of the choices that participants ultimately made. Study data were collected and managed using REDCap electronic data capture tools hosted at Northwestern University.37 After surveys were completed, the lead author contacted each site representative via phone or email to confirm and clarify responses. This method of cross-site comparison has been utilized in multiple prior consortium analyses.38,39
RESULTS
An overview of the recruitment methods for all ten eMERGE III sites is described in Table 1. Two sites had pediatric populations (CCHMC, CHOP), while the remaining sites had only adult participants. Most sites had multiple study cohorts, resulting in a total of 17 cohorts across the Network. Study cohorts varied based on indication for testing, including population and/or phenotype enrichment, and whether retrospective or prospective samples were sent for testing.
Five sites did not offer their participants choices at the time of consent about the types of results they wanted to receive. MMC initially consented their participants to receive only cancer risk-related results; however, participants found to be positive for other conditions on the eMERGE consensus list were contacted with the option to reconsent and receive those additional results. Of the sites that did not offer choices at any time point, two (GE, PHC) specified that their samples were retrospectively collected from biorepositories and participants were already consented to receive all actionable results, one site (VUMC) indicated that the return of all results was necessary for their study design, and one site (KPWA/UW) specified that choices were not considered in their study design.
Five sites offered their participants choices at the time of study or biobank consent about the types of results they wanted to receive. Three of these sites (CCHMC, CHOP, MC) offered all of their study participants choices, while two (CU, NU) offered choices to only specific cohorts. NU randomized a subset of their participants to either receive all results or be offered a choice of what types of results to receive. Three cohorts were recruited without a specific disease indication. Four cohorts included at least some participants who were recruited to the study because they were at risk for, or were suspected to have, a genetic condition. At MC, both disease indication–based cohorts received primary results by study design and could choose to learn results for the remaining genes on the consensus panel. At NU and CHOP, participants who were recruited based on disease indication were given choices related to all of the results, including potential primary findings.
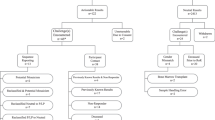
Of the five sites that offered participants choices, each developed unique categories of choices (Fig. S1). All had categories based on whether a condition was actionable, though the definition of actionability varied. Some used the availability of treatments to define actionability, including if the condition was completely treatable or if treatments were only partially effective, while other sites distinguished conditions that had screening or prevention options. Several sites categorized results based on nondisease risk (e.g., autosomal recessive carrier status, pharmacogenomic) and broad phenotypes (e.g., cancer risk, heart disease, behavior and neurocognitive conditions). The pediatric sites also categorized results based on age of onset. Furthermore, two sites (CCHMC, CU) offered participants the option to make granular choices about individual conditions. For these sites, participants were given a list of conditions based on their categorical choices. Participants could then choose to include or exclude specific conditions on the list. MC and CHOP offered participants options based on actionability, but the other sites used multiple different types of categories for their participant options.
The pediatric sites had additional unique selection protocols. CCHMC had two cohorts: a prospective cohort with adolescent/parent dyads (CCHMC-adol) in which decisions about the type of results the adolescent would receive were first made independently and then together by the adolescent and parent,26 as well as a retrospective biobank cohort (CCHMC-bio) in which parents made the selections about the types of results to learn about their child (age less than 18 years). While participants in CCHMC-adol were given the option to learn about adult-onset conditions, genes associated with adult-onset conditions were not analyzed for CCHMC-bio. At CHOP, parents made the selections for their children under 18 years old about the types of results to receive. The biobank samples submitted by CHOP were analyzed for genes associated with adult-onset conditions; however, site investigators deferred offering biobank participants the opportunity to learn such results until after they turned 18 years of age, reconsented to participate in the biobank, and agreed to be recontacted to learn the results.
Sites that offered choices referenced different sources to create their categories, including patient focus groups (CCHMC, CU), previous experience (CU), literature (CCHMC, CHOP, NU), and requirements from the site’s IRBs (CCHMC, CHOP). The sites that offered more than one category of choices assembled a group of stakeholders to sort genes and SNVs into their disease categories (CCHMC, CU, NU). Sites reported many challenges in this process. As the eMERGEseq panel was not finalized until well into the grant funding period, some sites created their categories without knowing exactly which genes would be tested (CHOP, CU, MC, NU). This resulted in a few hypothetical categories (e.g., dementia). NU and CCHMC reported challenges in handling genes that informed different phenotypes based on the type or location of pathogenic variants, thus resulting in some genes being sorted into more than one category. Finally, some results did not fit into a predetermined category (e.g., biological sex–gender identity discrepancies). Table 2 and Table S1 summarize the sites’ assigned categories for the genes and SNVs that were approved by their IRBs for potential return.
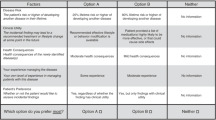
How the sites presented choices to participants varied. Sites either asked participants to check a box to indicate their choice to receive a specific category of result (CCHMC, CU, MC) or to indicate yes or no next to their options (CHOP, NU). Participants completed their selections on paper (CCHMC, CU, MC, NU, CHOP) or electronically (CU, NU). Sites used different language to describe what participants might learn: “information related to…” (NU), “results” (Mayo, CHOP), “gene change” (CCHMC-bio), “condition” (CCHMC-adol), and “genetic mutations” (CU). Some sites (CU, CCHMC-adol) provided definitions or additional information about terms used to describe the different results options, such as preventability, treatability, and carrier status. Finally, at some sites (CU, CCHMC-bio, MC), participants consented to the study and made their choices remotely with the option to speak with a study research assistant trained in genetics or make an appointment with the study genetic counselor. At other sites (NU, CCHMC-adol, CHOP, MC) the selections were made in the presence of a study staff member or a genetic counselor.
The selections that participants made are compiled in Table 3 and Table S2. In total, 4664 (18.6%) eMERGE III participants were offered choices. Most (94.5%, 4407/4664) elected to receive all results, while a portion (5.5%, 257/4664) opted to exclude some or all results. Sites that offered more choices than actionability alone had higher proportions of participants that excluded results for disease risk: 19.6% at CCHMC-adol, 18.2% at CU and 9.2% at NU, versus 1.8% at MC and 0% at CHOP. After viewing the list of conditions they would and would not learn based on categorical choices, 24 (14.7%) CCHMC adolescent–parent dyads made granular choices. At CU, 68% (40/59) of those who excluded any results made granular selections while 32% (19/59) made categorical choices only to exclude conditions. There were no apparent trends or patterns as to the types of granular selections that were made. All sites that offered choices allowed their participants to change their selections after consenting, with the exception of the CCHMC-bio cohort. For most sites, the ability to change their selections was deliberately reviewed and discussed prior to result disclosure (CCHMC-adol, CHOP, CU, MC). At NU, the option was available but not openly discussed. Few participants (0.5%, 21/4645) changed their selections after enrollment, with most (20/21) changing their selections after the ability was deliberately reviewed and discussed.
DISCUSSION
We present the largest reported study of individuals prospectively offered options about the types of genomic results they wanted to learn before they underwent testing. Although each site differed in the choices they offered participants, our collective results demonstrate that when provided with options, a subset of participants choose to learn some but not all potential results. Moreover, when offered the ability to make granular choices about individual conditions, some participants elect to do so. Similar to our results from a research setting, others have found that up to 18.4% of probands choose to learn some but not all of offered secondary findings when consenting for clinical sequencing.40
One unique strength of our study is that most of the sites that offered choices provided participants the ability to change their selections after their choices were made. Others have recommended a process that offers individuals the option to change their choices at a later date.34 We are unaware of any other studies that have actually given this option to participants who are undergoing sequencing. Our data show that some participants do change their minds. This finding provides empirical evidence supporting earlier recommendations, and the varied approaches used by eMERGE sites provide alternatives to consider when implementing an option to change choices. Additionally, our data show that actively offering participants the option to change their choices results in more selection changes than a passive approach in which the option is available but not directly discussed with the participant. Future studies of the motivations behind the change of choice would also inform best practices for offering choices.
Our study was a natural experiment that presents many different strategies for providing individuals with options to learn their results. Our findings provide considerations for clinical practice, as well as the protocols of other research consortiums and large-scale studies. Superiority of one method over another cannot be determined from our data. Our results support that a method for providing choices will need to consider the population and environments within which choices are made and results eventually returned. It is important to provide meaningful genomic result choices in a constantly changing field where anticipating all possible results is currently impossible. Yet, the feasibility of sorting genes and conditions neatly into distinct categories is a moving target as treatment options change, our understanding of the natural history of diseases improves, penetrance estimates are refined, and we identify more genes that contribute to more than one phenotype. Additionally, there is a need to solicit greater involvement from the community who will be making these choices and therefore will be affected by them. Only two of the sites involved participant stakeholders to inform their results categories. Ideally, how categories are determined and genes are sorted would be a fluid process to account for advances in our understanding of genes and phenotypes, and our growing understanding of how individuals make choices.
This study had limitations. While our protocols and findings can help to inform clinical practice, the selections that participants made cannot be generalized to a patient population undergoing testing for a clinical diagnosis, as many of our participants had no indication for testing. Additionally, we did not attempt to understand why participants made the selections that they did or why some participants changed their choices. We were unable to form any conclusion about the different choices made across cohorts because of the heterogeneity of the cohorts, choices offered and study protocols. Future research should interview participants about the choices they make, reasons for changing choices, and the factors that influence their selections. Satisfaction with choices after results have been returned is another area of future research. Finally, while our study utilizes an accepted method to present consortium data and limitations, the variability in the cohorts, study protocols, and choices offered did not allow us to assess for the significance of covariables such as age, race, gender, education, geographic location, or disease status. While our approach more accurately reflects real-world implementation, to determine the impact of cohort differences, future consortium studies should develop a consensus approach prior to study implementation for which choices are offered, how genes and variants are categorized, and the timing of when participants are offered the opportunity to change their choices.
References
Daack-Hirsch S, Driessnack M, Hanish A, et al. ‘Information is information’: a public perspective on incidental findings in clinical and research genome-based testing. Clin Genet. 2013;84:11–18.
Facio FM, Eidem H, Fisher T, et al. Intentions to receive individual results from whole-genome sequencing among participants in the ClinSeq study. Eur J Hum Genet. 2013;21:261–265.
Jelsig AM, Qvist N, Brusgaard K, Ousager LB. Research participants in NGS studies want to know about incidental findings. Eur J Hum Genet. 2015;23:1423–1426.
Shahmirzadi L, Chao EC, Palmaer E, Parra MC, Tang S, Gonzalez KD. Patient decisions for disclosure of secondary findings among the first 200 individuals undergoing clinical diagnostic exome sequencing. Genet Med. 2014;16:395–399.
Yu JH, Crouch J, Jamal SM, Tabor HK, Bamshad MJ. Attitudes of African Americans toward return of results from exome and whole genome sequencing. Am J Med Genet A. 2013;161a:1064–1072.
Yu JH, Crouch J, Jamal SM, Bamshad MJ, Tabor HK. Attitudes of non-African American focus group participants toward return of results from exome and whole genome sequencing. Am J Med Genet A. 2014;164a:2153–2160.
Clift KE, Halverson CM, Fiksdal AS, Kumbamu A, Sharp RR, McCormick JB. Patients’ views on incidental findings from clinical exome sequencing. Appl Transl Genom. 2015;4:38–43.
Fernandez CV, Bouffet E, Malkin D, et al. Attitudes of parents toward the return of targeted and incidental genomic research findings in children. Genet Med.2014;16:633–640.
Wynn J, Martinez J, Bulafka J, et al. Impact of receiving secondary results from genomic research: a 12-month longitudinal study. J Genet Couns. 2018;27:709–722.
Fiallos K, Applegate C, Mathews DJ, Bollinger J, Bergner AL, James CA. Choices for return of primary and secondary genomic research results of 790 members of families with Mendelian disease. Eur J Hum Genet. 2017;25:530–537.
Gray SW, Park ER, Najita J, et al. Oncologists’ and cancer patients’ views on whole-exome sequencing and incidental findings: results from the CanSeq study. Genet Med. 2016;18:1011–1019.
Ross LF, Saal HM, David KL, Anderson RR. Technical report: ethical and policy issues in genetic testing and screening of children. Genet Med. 2013;15:234–245.
Abdul-Karim R, Berkman BE, Wendler D, et al. Disclosure of incidental findings from next-generation sequencing in pediatric genomic research. Pediatrics.2013;131:564–571.
Botkin JR, Belmont JW, Berg JS, et al. Points to consider: ethical, legal, and psychosocial implications of genetic testing in children and adolescents. Am J Hum Genet. 2015;97:6–21.
Committee on Bioethics, Committee on Genetics, American College of Medical Genetics, et al. Ethical and policy issues in genetic testing and screening of children. Pediatrics. 2013;131:620–622.
Kleiderman E, Knoppers BM, Fernandez CV, et al. Returning incidental findings from genetic research to children: views of parents of children affected by rare diseases. J Med Ethics. 2014;40:691–696.
Sapp JC, Dong D, Stark C, et al. Parental attitudes, values, and beliefs toward the return of results from exome sequencing in children. Clin Genet.2014;85:120–126.
Myers MF, Martin LJ, Prows CA. Adolescents’ and parents’ genomic testing decisions: associations with age, race, and sex. J Adolesc Health. 2020;66:288–295.
Pervola J, Myers MF, McGowan ML, Prows CA. Giving adolescents a voice: the types of genetic information adolescents choose to learn and why. Genet Med. 2019;21:965–971.
Bush LW, Bartoshesky LE, David KL, Wilfond B, Williams JL, Holm IA. Pediatric clinical exome/genome sequencing and the engagement process: encouraging active conversation with the older child and adolescent: points to consider—a statement of the American College of Medical Genetics and Genomics (ACMG). Genet Med. 2018;20:692–694.
Berg JS, Amendola LM, Eng C, et al. Processes and preliminary outputs for identification of actionable genes as incidental findings in genomic sequence data in the Clinical Sequencing Exploratory Research Consortium. Genet Med. 2013;15:860–867.
Berg JS, Foreman AK, O’Daniel JM, et al. A semiquantitative metric for evaluating clinical actionability of incidental or secondary findings from genome-scale sequencing. Genet Med. 2016;18:467–475.
Kalia SS, Adelman K, Bale SJ, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med. 2017;19:249–255.
Bacon PL, Harris ED, Ziniel SI, et al. The development of a preference-setting model for the return of individual genomic research results. J Empir Res Hum Res Ethics. 2015;10:107–120.
Ackerman SL, Koenig BA. Understanding variations in secondary findings reporting practices across U.S. genome sequencing laboratories. AJOB Empir Bioeth. 2018;9:48–57.
Jarvik GP, Amendola LM, Berg JS, et al. Return of genomic results to research participants: the floor, the ceiling, and the choices in between. Am J Hum Genet. 2014;94:818–826.
Bombard Y, Clausen M, Mighton C, et al. The Genomics ADvISER: development and usability testing of a decision aid for the selection of incidental sequencing results. Eur J Hum Genet. 2018;26:984–995.
Berg JS, Khoury MJ, Evans JP. Deploying whole genome sequencing in clinical practice and public health: meeting the challenge one bin at a time. Genet Med. 2011;13:499–504.
Boardman F, Hale R. Responsibility, identity, and genomic sequencing: a comparison of published recommendations and patient perspectives on accepting or declining incidental findings. Mol Genet Genomic Med. 2018;6:1079–1096.
Brothers KB, East KM, Kelley WV, et al. Eliciting preferences on secondary findings: the Preferences Instrument for Genomic Secondary Results. Genet Med. 2017;19:337–344.
Wynn J, Martinez J, Duong J, et al. Research participants’ preferences for hypothetical secondary results from genomic research. J Genet Couns. 2017;26:841–851.
Jamal L, Robinson JO, Christensen KD, et al. When bins blur: patient perspectives on categories of results from clinical whole genome sequencing. AJOB Empir Bioeth.2017;8:82–88.
Thorogood A, Dalpe G, Knoppers BM. Return of individual genomic research results: are laws and policies keeping step? Eur J Hum Genet. 2019;27:535–546.
Mackley MP, Fletcher B, Parker M, Watkins H, Ormondroyd E. Stakeholder views on secondary findings in whole-genome and whole-exome sequencing: a systematic review of quantitative and qualitative studies. Genet Med. 2017;19:283–293.
eMERGE Consortium. Harmonizing clinical sequencing and interpretation for the eMERGE III Network. Am J Hum Genet. 2019;105:588–605.
Wiesner GL, Kulchak Rahm A, Appelbaum P, et al. Returning results in the genomic era: initial experiences of the eMERGE Network. J Pers Med. 2020;10:E30.
Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)-a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42:377–381.
Porter KM, Kauffman TL, Koenig BA, et al. Approaches to carrier testing and results disclosure in translational genomics research: The clinical sequencing exploratory research consortium experience. Mol Genet Genomic Med. 2018;6:898–909.
Wolf SM, Amendola LM, Berg JS, et al. Navigating the research-clinical interface in genomic medicine: analysis from the CSER Consortium. Genet Med. 2018;20:545–553.
Bishop CL, Strong KA, Dimmock DP. Choices of incidental findings of individuals undergoing genome wide sequencing, a single center’s experience. Clin Genet. 2017;91:137–140.
Acknowledgements
This phase of the eMERGE Network was initiated and funded by the National Human Genome Research Institute (NHGRI) through the following grants: U01HG008657 (Group Health Cooperative/University of Washington); U01HG008685 (Brigham and Women’s Hospital); U01HG008672 (Vanderbilt University Medical Center); U01HG008666 (Cincinnati Children’s Hospital Medical Center); U01HG006379 (Mayo Clinic); U01HG008679 (Geisinger Clinic); U01HG008680 (Columbia University Irving Medical Center); U01HG008684 (Children’s Hospital of Philadelphia); U01HG008673 (Northwestern University); U01HG008701 (Vanderbilt University Medical Center serving as the Coordinating Center); U01HG008676 (Partners Healthcare/Broad Institute); U54MD007593-10 (Meharry Medical College); and U01HG008664 (Baylor College of Medicine).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure
W.K.C. serves on the Scientific Advisory Board of Regeneron Genetics Center. K.M. is a consultant for Novartis, as well as a coinventor and consultant for Hive Networks, Inc. The other authors declare no conflicts of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary in formation
Rights and permissions
About this article
Cite this article
Hoell, C., Wynn, J., Rasmussen, L.V. et al. Participant choices for return of genomic results in the eMERGE Network. Genet Med 22, 1821–1829 (2020). https://doi.org/10.1038/s41436-020-0905-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-020-0905-3
Keywords
This article is cited by
-
Returning individual genomic results to population-based cohort study participants with BRCA1/2 pathogenic variants
Breast Cancer (2023)
-
An spanish study of secondary findings in families affected with mendelian disorders: choices, prevalence and family history
European Journal of Human Genetics (2023)
-
Public interest in unexpected genomic findings: a survey study identifying aspects of sequencing attitudes that influence preferences
Journal of Community Genetics (2022)