Abstract
Purpose
To identify novel genes associated with intellectual disability (ID) in four unrelated families.
Methods
Here, through exome sequencing and international collaboration, we report eight individuals from four unrelated families of diverse geographic origin with biallelic loss-of-function variants in UBE4A.
Results
Eight evaluated individuals presented with syndromic intellectual disability and global developmental delay. Other clinical features included hypotonia, short stature, seizures, and behavior disorder. Characteristic features were appreciated in some individuals but not all; in some cases, features became more apparent with age. We demonstrated that UBE4A loss-of-function variants reduced RNA expression and protein levels in clinical samples. Mice generated to mimic patient-specific Ube4a loss-of-function variant exhibited muscular and neurological/behavioral abnormalities, some of which are suggestive of the clinical abnormalities seen in the affected individuals.
Conclusion
These data indicate that biallelic loss-of-function variants in UBE4A cause a novel intellectual disability syndrome, suggesting that UBE4A enzyme activity is required for normal development and neurological function.
Similar content being viewed by others
INTRODUCTION
Intellectual disability (ID) occurs in 1–3% of the general population and is defined by limitations in intellectual functioning and adaptive behavior.1,2 ID is characterized by significant genetic heterogeneity with over 1,000 genes implicated to date, while thousands of genes likely remain to be identified.3,4,5,6 Next-generation sequencing technologies and collaborative research networks have greatly advanced the identification and characterization of ID-related genes. In most families, ID is sporadic, typically due to a de novo pathogenic variant. However, there are likely many rare Mendelian genetic causes of ID associated with autosomal recessive inheritance, most of which have not yet been described.3 Genes involved in protein ubiquitination play a significant role in ID and neurodevelopmental disorders (NDDs) and remain an important pathway for discovery of ID-related genes.
A careful balance of protein synthesis and degradation is required to maintain cellular protein steady-state and proper function. Protein ubiquitination is a highly conserved, post-translational modification that labels proteins for diverse functions within the cell as well as misfolded proteins for degradation via the ubiquitin–proteasome system (UPS).7,8 Overall, the ubiquitin–proteasome pathway has two critical steps: (1) covalent attachment of ubiquitin to the targeted protein substrate followed by several ubiquitin molecules creating a chain assembly, and (2) degradation of the ubiquitylated protein by the proteasome complex. To do this, the UPS requires an ubiquitin-activating enzyme (E1), an ubiquitin conjugating enzyme (E2), and a substrate-specific ubiquitin-protein isopeptide ligase (E3) to covalently attach the 76–amino acid protein ubiquitin to proteins targeted for degradation. This multistep process plays an important role in protein degradation including that of many short-lived regulatory proteins, such as those that contribute to the cell cycle and cellular signaling in response to DNA repair, environmental stress, secretion, and morphogenesis.9 U-box proteins, including UBE4A, were initially classified as a fourth class of ubiquitin molecule, ubiquitination protein ligase (E4) and thought to promote polyubiquination and efficient ubiquitin chain assembly.7,8 Additional studies demonstrated that U-box proteins can uniquely function as both E3 and E4 ligases.10,11
Impairment of ubiquitination can cause ID and other neurological conditions due to multiple mechanisms, including the decline of the cellular proteolytic capacity, accumulation of misfolded protein, and reduction of effective protein production.12,13,14 Several genes involved in the UPS-mediated degradation pathway are known to cause syndromic NDD including UBE3A (Angelman syndrome, [MIM 105830]),12 and more recently described UBE2A (MIM 300860)13 and UBE3B (MIM 244450).14 An extensive list of many other E2, E3 ligases, and deubiquitinating enzymes have been described in ID and NDD (e.g., HUWE1, HERC2, HECW2, UBR1, USP7), suggesting that variants in ubiquitin-related genes could explain some unsolved ID cases.15 However, our understanding of UBE4A function in humans and its implication in ID is sparse. Biallelic UBE4A loss-of-function (LoF) variants were previously proposed as a candidate ID gene in a single family, also reported here (family C), but have not otherwise been described.5
Through the GeneMatcher platform16 and professional conversations with colleagues, we collaboratively assembled a cohort of eight individuals in four families, all with syndromic ID and global developmental delay. All affected individuals had homozygous LoF UBE4A variants. Functional studies presented further evidence for pathogenicity, showing degradation of UBE4A in vitro. Mice engineered with a pathogenic variant of Ube4a exhibited ocular abnormalities, musculoskeletal weakness, and behavioral hypersensitivity, which were also, to some degree, observed in our cases. Herein, we report eight individuals with homozygous UBE4A LoF variants, with a consistent phenotype and recognizable features, in characterizing the novel UBE4A-related intellectual disability syndrome.
MATERIALS AND METHODS
Human subjects and genetic analysis
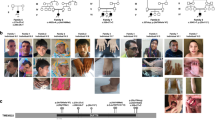
Exome sequencing was performed in individual A-1 (Fig. 1). After confirmation that the candidate variant in UBE4A segregated with disease in family A, additional individuals with UBE4A variants were identified with the use of GeneMatcher16 and colleague correspondence. This resulted in identification of eight individuals in four unrelated families located in Brazil, the United States, Saudi Arabia, and Germany (Table 1; Table S1). In each additional family, candidate UBE4A variant was detected by trio exome sequencing. Family C was reported in a previously published research study of 337 individuals with unresolved intellectual disability; UBE4A was noted as a candidate gene.5 Given that each subject received a nondiagnostic clinical exome result, subjects were subsequently enrolled in research for additional molecular and functional investigation.
(a) Clinical pictures from families A–D. (b) Pedigree of four studied families and segregation of the UBE4A pathogenic variants. Pathogenic variants are listed above each pedigree. Solid symbols represent affected individuals. −/− = homozygous pathogenic variant; +/− = heterozygous. *Samples were genotyped but data not shown. (c) Schematic overview of UBE4A (NM_004788.3) that includes domain organization, exon boundaries, and pathogenic variants detected in this study.
Exome and Sanger sequencing
Trio exome sequencing (ES) was independently performed in each family. ES was performed for the proband in family A and as a trio in families B, C, and D. Technical details have been previously described for family C.5 In brief, sample library was prepared using Nextera Extension Exome Rapid-Capture kit (Illumina, San Diego, CA, USA) for family A; Agilent SureSelect Clinical Research Exome kit mixed with an enhanced capture protocol for family B; Ion Proton AmpliSeq library using Exome Primer Pools and AmpliSeq HiFi mix (Thermo Fisher, Carlsbad, CA, USA) for family C; and xGEN (IDT, Coralville, IA, USA) with subsequent TruSeq DNA Library Prep for Enrichment (IDT Exome, Illumina) for family D. Exome libraries were run on Illumina HiSeq2500 (Illumina) for families A and B, Ion Proton instrument (Thermo Fisher) for family C, and NextSeq (Illumina) for family D. Sequencing reads were aligned to the reference genome GRCh37/hg19 and variants were filtered according to established pipelines (Supplemental Material). In each family, candidate variants were assessed for segregation analysis by polymerase chain reaction (PCR) followed by Sanger sequencing, using primers described in Table S2.
Fibroblast and LCL cell culture
Functional studies were performed for families A and D. Skin biopsy was obtained on patients A-1, A-2 by standard procedure. Lymphoblastoid cell lines (LCLs) were established by Epstein–Barr virus (EBV) transformation of leukocytes from peripheral blood samples of individual D-1. Fibroblasts and LCLs from unrelated healthy individuals were used as controls. Fibroblasts were treated overnight in DMEM/F12 media with dispase (1 U/mL, Stemcell Technologies), then plated in a dish with DMEM high-glucose fibroblast media (Thermo Fisher, Waltham, MA, USA) supplemented with 10% fetal bovine serum (Thermo Fisher), 1% nonessential amino acids (NEAA; Merck, Darmstadt, Germany), and 1% penicillin–streptomycin (Sigma-Aldrich, St. Louis, MO, USA).
Peripheral blood mononuclear cells (PBMCs) were obtained from sodium-heparin blood of patient D-1 by dilution with complete medium consisting of RPMI 1640 Glutamax supplemented with 15% FCS, 1% penicillin–streptomycin (Gibco, Thermo Fisher) and gradient centrifugation with Ficoll (Merck). PBMCs were washed twice with complete medium and resuspended with EBV (B95-8) supernatant (50%) supplemented with complete medium (48.5%) and PHL-A buffer (1.5%, Biochrom, Merck). After incubation with transformation medium for 2–3 days, immortalized cells were cultured in complete medium.
Expression analysis
RNA was extracted from fibroblasts from families A (A-1, A-2) and D (D-1) using RNeasy mini kit (Qiagen, Hilden, Germany). Total RNA (2 µg/µL) was reverse-transcribed using oligo(dT) primers and SuperScript™ III First-strand Synthesis System (Thermo Fisher). Primers for reverse transcription quantitative PCR (RT-qPCR) are described in Table S2. RT-qPCR was performed using the Applied Biosystems® 7500 Fast Real-time PCR System and gene expression was calculated using the 2−ΔΔCT method.17
Protein extraction and western blot analysis were performed using standard protocols using A-1 and A-2 samples. Whole-cell lysates were obtained by lysing fibroblasts with RIPA buffer (150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris-HCl, pH 8) containing protease inhibitor cocktail (Sigma-Aldrich). Protein concentrations were determined using Pierce™ BCA Protein Assay Kit (Thermo Fisher) according to the manufacturer’s protocol. Proteins were separated using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto nitrocellulose membranes. Primary antibodies used were anti-UBE4A EPR7332 (Abcam, ab137074, rabbit) and anti-beta actin [Ac-15] HRP (Abcam, ab49900, mouse). Anti-UBE4A (1:1000) and anti-beta actin (1:50000) were diluted in TBST buffer (20 mM Tris, pH 7.5; NaCl 150 mM; 0,1% Tween 20) and incubated for 1 hour at room temperature. Secondary immunodecoration was performed using horseradish peroxidase (HRP)-conjugated antirabbit (Cell Signaling). The secondary antibody was diluted (1:10000) in TBST buffer and incubated for 1 hour at room temperature.
Design and targeting using CRISPR/Cas9
The online tool CHOPCHOP was used to assess optimal guide RNA (gRNA) candidates and avoid greater than two mismatches anywhere in the genome to prevent off-target endonuclease activity.18 The gRNA sequence (Table S2) was selected and synthesized as CRISPR RNA (crRNA) for use as an Alt-R™ two-part guide system with trans-activating crRNA (tracrRNA) and crRNA provided separately (Integrated DNA Technologies, Coralville IA). An ssODN repair template was designed with an engineered two-nucleotide base pair deletion resulting in the C425Ffs* modification mimicking the pathogenic variant present in family B (p.Cys413Phefs*55) and with the addition of silent mutations to protect against recleavage. Template genome homology arms consisted of an offset method19 synthesized as an ultramer (Integrated DNA Technologies, Coralville IA). Knock-in allele CRISPR preparation, zygote treatment, and electroporation were performed following the CRISPR-EZ method with a final concentration of 8 uM RNP; 10 uM ssODN.20 After embryo transfer and birth of pups, genomic DNA was extracted from tail snips using Agencourt® DNAdvance™ magnetic beads (Beckman–Coulter, A48706, Brea, CA, USA) on an automated and LIMS tracked Microlab STAR (Hamilton Robotics, Reno, NV, USA). Using a quantitative relative Ct method, purified DNA samples were screened for the presence of the engineered sequence modification and quantified adjacent to an endogenous Tcrd reference in multiplex.21 Positive samples were PCR amplified with locus specific primers (Table S2) using GoTaq®G2 kit following manufacturer’s recommendation (Promega, M7433, Madison, WI, USA). The expected amplicon size (511 bp) was gel purified using QIAquick Gel extraction kit following manufacturer’s instruction (Qiagen, 28506, Hilden, Germany) and subjected to Sanger sequencing to confirm integration of the repair template by homology directed repair (HDR). HDR positive mice were backcrossed to generate cohorts of animals for phenotyping.
Mouse phenotyping
Male and female cohorts (n = 3 each) of mice homozygous for the C425F frameshift (fs) inducing mutation (Ube4aC425Ffs/Mbp) made on an inbred C57BL/6N genetic background and age and sex-matched wild-type control (C57BL/6N) mice underwent a series of in vivo analytical measures and tests targeted to assess the presence (or absence) of comparable phenotypes observed during the clinical presentation of family B. Variable observations were made at specific ages over a 16-week period and included weekly body weights, neuromuscular function (grip strength), behavior and cognition evaluation (open field, prepulse inhibition, social novelty, and marble burying), and a complete (direct and indirect) ophthalmological examination.
Statistical analysis
Statistical analysis was performed using GraphPad Prism (San Diego, CA, USA). Two-way analysis of variance (ANOVA), t-test, and Mann–Whitney were applied. Descriptive statistics of mouse phenotype data were analyzed using SAS (Version 9.14). Single parameter data (grip strength, social novelty, marbles burying, and open field) were analyzed using Wilcoxon nonparametric exact test.
RESULTS
Clinical phenotype
Neurodevelopment is significantly impaired in all eight individuals and was the primary indication for genetic evaluation in infancy (Table 1). Individuals are globally delayed with speech limited to single words or short sentences and the majority (n = 6/8) being nonambulatory. Individuals B-3 and C-1 lost the ability to walk after taking independent steps at 5 years and 3 years respectively, largely considered secondary to dyspraxia with some contribution from hypotonia (Table S1). No other developmental regression is noted. Other common neurologic features included seizures (n = 5/8) and behavioral abnormalities (n = 6/8) such as aggression, attention deficit–hyperactivity disorder (ADHD), and autistic behavior (Table 1). When present, seizure type was tonic–clonic (n = 5/5) with onset ranging between 10 months and 5 years. Brain magnetic resonance image (MRI) (1.5 T equipment) was normal or nonspecific in most individuals (n = 5/8). Although birth parameters were reported typical, short stature by mid-childhood was common (n = 4/6). Obesity and a compact body habitus (which we defined as short stature, decreased muscle mass, and increased body fat) was noted in families B and C. In most individuals, mild dysmorphic features were noted (Fig. 1a) including prominent nose with a bulbous tip (n = 8/8), dental anomalies such as large or widely spaced teeth (n = 5/7), deep set eyes (n = 6/8), upslanted palpebral fissures (n = 3/8), small, square, puffy hands and short feet (n = 5/6) (Figure S1). Adolescent-onset cataracts were noted in family B (n = 2/8, Figure S2). A case report for each subject is included in the Supplemental Material.
Molecular results and spectrum of pathogenic variants
Exome sequencing was independently performed for each family. The LoF variants in UBE4A detected in this study comprise two frameshift variants (NM_004788.3:c.1238_1239del; NP_004779.2:p.[Cys413PhefsTer55] and NM_004788.3:c.1206_1207del; NP_004779.2:p.[Lys403ThrfsTer65]) and two nonsense variants (NM_004788.3:c.384G>A; NP_004779.2:p.[Trp128Ter] and NM_004788.3:c.631C>T; NP_004779.2:p.[Arg211Ter]) (Fig. 1b, c). Variants were identified by prioritizing variants that were rare (minor allele frequency [MAF] <0.01) in population databases, inherited in a recessive manner, resulted in LoF, and were in genes having no LoF variants in homozygosity based on gnomAD (v2.1.1). All identified variants are predicted to shorten the UBE4A messenger RNA (mRNA) transcript, resulting in loss of the highly conserved U-box protein functional domain and likely subject to nonsense-mediated mRNA decay. Sanger sequencing was performed to confirm all variants in the homozygous state in affected individuals (Fig. 1b). All parents were heterozygous for respective variants as expected for an autosomal recessive disorder. None of the tested unaffected siblings were homozygous for respective variants.
UBE4A LoF variants result in decreased RNA expression and protein abundance
All UBE4A variants described here are predicted to be LoF. Functional studies to confirm LoF were performed for family A and family D. RT-qPCR using complementary DNA (cDNA) reverse-transcribed from available samples revealed downregulation of UBE4A transcripts up to 80% in affected fibroblasts samples (A-1 and A-2) when compared with controls; in LCLs (D-1), UBE4A transcripts were mostly undetected (Fig. 2a). Western blot showed absent UBE4A protein in samples from family A (Fig. 2b).
(a) Reverse transcription quantitative polymerase chain reaction (RT-qPCR) results showing downregulation of UBE4A transcripts in fibroblasts from family A and lymphoblastoid cell lines (LCLs) from family D (black dots), compared with respective controls (white triangles). (b) Western blot results using antibody against UBE4A show absence of this protein in samples from A-1 and A-2.
Ube4a C425Ffs* homozygote mice showed phenotypic differences and ocular abnormalities
Next, using CRISPR/cas9 genome editing system, we generated a mouse line mimicking the LoF variant detected in family B (Ube4aC425Ffs*). Behavior, cognition, and neuromuscular strength and coordination measures were evaluated on a small number of mutant male and female mice to quickly assess the likelihood that the Ube4aC425Ffs* mutation was causative of the phenotype observed in family B. Assessments/measures and time points are shown in Figure S3, and the descriptive statistics for all test results are presented in Table S3.
Eye examination of mice revealed that homozygous male and female Ube4aC425Ffs* mice (n = 5/6) developed bilateral early cataracts described as mild punctate lens opacities, noted at 8 weeks postnatal age (Fig. 3). However, mild punctate lens opacities lesions were also seen in control animals to lesser extent; it is unclear if the Ube4aC425Ffs* variation causes lens opacity in mice. Noteworthy, homozygous Ube4aC425Ffs* mice have increased retinal dysplasia at 16 weeks postnatal compared with controls (Fig. 3d).
For neuromuscular functions, abnormal limb grasp was noted in one of three male and female Ube4aC425Ffs* mice (SHIRPA, 6 weeks of age; data not shown). A reduction of forelimb grip strength was observed in both male and female Ube4aC425Ffs* mice (12 weeks of age) compared with controls (Fig. 4a), reaching statistical significance when pooling male and female mice data (Fig. 4b). A similar observation occurred with all limbs grip strength (Fig. 4c), the difference between mutants and controls becoming significant after pooling (Fig. 4d).
Results are mean ± standard error of n = 3 males and n = 3 females for Ube4aC425Ffs*, and n = 17 and n = 14 for WT, respectively. (a–d) Grip strength of forelimbs and all limbs used to assess neuromuscular function, normalized for body weight. (a) Slight sex differences were observed in forelimb grip parameters, although not significant. Females Ube4a have lower grip strength than WT females. (b) When sex/condition data are pooled, we observed a significantly smaller forelimbs grip force strength (Wilcoxon scores). (c,d) All limbs grip strength is decreased in Ube4aC425Ffs* males and females and significantly lower when sex/condition data are pooled together (Wilcoxon scores). (e) Results show no sex differences in startle parameters (Ube4a/WT). (f) PPIG is significantly lower in females/males Ube4aC425Ffs compared with WT (Wilcoxon score) and the difference appears to be caused by a much attenuated PPI at the lower db (PPI1).
In response to the acoustic startle test (10 weeks of age), Ube4aC425Ffs* mice tended to show less attenuation of startle in response to a prepulse stimulus at all three levels of decibels tested (Fig. 4e, f). In addition, we noted that two females show compromised auditory function.
Small differences were noted in the responses to the open field, marble burying, and social novelty recognition between Ube4aC425Ffs* mice compared with controls (Figure S4; Supplemental Material). Other measurements (e.g., heart and body weight) were also evaluated and described in the Supplemental Material (Figure S5). Taken together, the homozygous Ube4aC425Ffs* mice recapitulate some traits seen in our cases, reinforcing that this gene is associated with UBE4A-syndrome phenotype.
DISCUSSION
Herein, we describe four families with homozygous, nonrecurrent, LoF variants in UBE4A, causing syndromic intellectual disability. The families were of diverse geographic origin, from Brazil, Guatemala, Saudi Arabia, and Iraq. All affected individuals had intellectual disability and global developmental delay. Severe, infantile hypotonia was noted in all individuals except D-1. Half of the affected individuals never took independent steps and two individuals lost the ability to walk before 5 years old. Speech was limited to single words or simple phrases in all affected individuals. Clinical features were variable but frequently included short stature (n = 5/8 individuals), behavior abnormalities (n = 6/8), and seizures (n = 5/8). In this cohort, only B-2 and B-3 had childhood-onset lamellar cataracts (onset at 13 and 12 years respectively). No other variants were detected that would explain cataracts in this family. Although cataracts were not appreciated in any other family, all other subjects were younger than the age of cataract onset in family B. The oldest individual (B-1) is 20 years and was followed at a single center since age 6 months. Neurologic examination was abnormal in infancy, which brought the children to medical attention. However, dysmorphic features developed with time in families A and B, becoming more apparent by middle childhood. Our cohort is small and relatively young; the identification of additional patients will be needed to clarify the natural history and phenotypic spectrum of the UBE4A-related disorder.
Observations in the induced mutant mouse model was intended as a pilot study to assess the likelihood that the homozygous UBE4A LoF variant in individuals from family B was likely pathogenic. For example, excessive marble burying is associated with repetitive, compulsive behaviors and anxiety, and the results of the two cognitive tests (acoustic startle and social novelty) are consistent with dysfunction in focused attention capacity and sociability. Our tests also revealed sexual dimorphism in some responses. These findings in Ube4aC425Ffs* mice support the causative and likely pathogenic nature of the same genetic variant in the human patient. Further, these results provide a compelling rationale for conducting further analysis and hypothesis testing in larger numbers of mice to confirm these findings statistically. Overall, these provide avenues of testing specific areas of behavior and cognition that can be supportive of the behavior anomalies appreciated in patients (n = 6/8), including aggressive behavior (n = 2/8) and autism spectrum disorder (n = 2/8). Hypotonia was a consistent feature appreciated in the majority (n = 7/8) of patients. Body mass index (BMI) ranged from 19.5 to 35, although the majority were overweight or obese, with BMI >25 (n = 5/6).
Although Ube4aC425Ffs* mouse showed ophthalmologic differences compared with control mice, including more severe retinal dysplasia, both Ube4aC425Ffs* mice and control mice developed cataracts with similar frequency and severity (Figure S3). Therefore, mouse studies did not resolve whether lenticular opacity is part of the UBE4A spectrum of disease. Notably, in humans, cataracts only presented in adolescence, and only in family B, after which the mice were modeled. It is unclear if cataracts are part of the UBE4A LoF phenotype, possibly with age-related penetrance or due to a separate etiology. Further ophthalmologic assessment would require additional human natural history study or, as control mice also developed cataracts, alternative mouse modeling.
The prototypic E4 enzyme, Ufd2 (ubiquitin fusion degradation), was first identified in Saccharomyces cerevisiae. It is defined by a U-box essential functional domain and highly conserved through evolution.9 Ufd2 was initially characterized as a proteolytic factor (E4) that catalyzes the elongation of pre-existing ubiquitin chains in collaboration with E1, E2, and E3 enzymes. Some studies suggested that Ufd2 (yeast) and its orthologue UBE4A participates in the UPS pathway, tagging misfolded proteins for proteasome degradation as a response to endoplasmic reticulum (ER) stress.9,22 Until recently, relatively little was known about Ufd2 human orthologues, UBE4A and UBE4B. Baranes-Bachar et al.23 demonstrated that UBE4A activity is required for DNA damage repair and the dynamic reorganization of proteins at the site of DNA damage. ER-related stress and/or impaired DNA damage repair may contribute to the observed phenotype and additional functional studies are needed to elucidate the underlying pathomechanism of disease.
Of note, UBE4A is located on 11q23.3, a locus commonly deleted in neuroblastoma and UBE4A has been demonstrated to play a role in targeted protein degradation in somatic cancer cells such as PCBP1 in thyroid cancer cells and SLAP in colorectal cancer cells.24,25,26 To date, no subjects have demonstrated cancer or a related phenotype; the oldest subject in our cohort is only age 20 years. Additional, longer natural history study may inform potential oncologic risks.
In summary, we present strong genetic evidence that LoF pathogenic variants in UBE4A cause syndromic autosomal recessive intellectual disability. The overlap of some phenotypes between Ube4aC425Ffs* mice and the families presented here further supports developing hypothesis driven testing to investigate a causative role for UBE4A. Given the clinical, molecular, and functional data presented here, we propose that the homozygous UBE4A LoF causes abnormal development and neurological dysfunction.
URLs
CHOPCHOP: http://chopchop.cbu.uib.no.
gnomAD: https://gnomad.broadinstitute.org/.
Data availability
Data and materials are available upon request.
References
Schalock, R. L., Borthwick-Duffy, S. A. & Bradley, M. et al. Intellectual Disability: Definition, Classification, and Systems of Supports 11th edn. (American Association on Intellectual and Developmental Disabilities, Washington DC, 2010).
Maulik, P. K., Mascarenhas, M. N., Mathers, C. D., Dua, T. & Saxena, S. Prevalence of intellectual disability: a meta-analysis of population-based studies. Res. Dev. Disabil. 32, 419–436 (2011).
Kochinke, K. et al. Systematic phenomics analysis deconvolutes genes mutated in intellectual disability into biologically coherent modules. Am. J. Hum. Genet. 98, 149–164 (2016).
Anazi, S. et al. Expanding the genetic heterogeneity of intellectual disability. Hum. Genet. 136, 1419–1429 (2017).
Anazi, S. et al. Clinical genomics expands the morbid genome of intellectual disability and offers a high diagnostic yield. Mol. Psychiatry 22, 615–624 (2017).
Monies, D. et al. Lessons learned from large-scale, first-tier clinical exome sequencing in a highly consanguineous population. Am. J. Hum. Genet. 104, 1182–1201 (2019).
Hershko, A. & Ciechanover, A. The ubiquitin system for protein degradation. Annu. Rev. Biochem. 61, 761–807 (1992).
Ravid, T. & Hochstrasser, M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat. Rev. Mol. Cell. Biol. 9, 679–690 (2008).
Koegl, M., Hoppe, T., Schlenker, S., Ulrich, H. D., Mayer, T. U. & Jentsch, S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 96, 635–644 (1999).
Tu, D., Li, W., Ye, Y. & Brunger, A. T. Structure and function of the yeast U-box-containing ubiquitin ligase Ufd2p. Proc. Natl. Acad. Sci. U S A 104, 15599–15606 (2007).
Hellerschmied, D. et al. UFD-2 is an adaptor-assisted E3 ligase targeting unfolded proteins. Nat Commun 9, 484 (2018).
Kishino, T., Lalande, M. & Wagstaff, J. UBE3A/E6-AP mutations cause Angelman syndrome [published correction appears in Nat Genet 1997 Apr;15(4):411]. Nat. Genet. 15, 70–73 (1997).
Nascimento, R. M., Otto, P. A., de Brouwer, A. P. & Vianna-Morgante, A. M. UBE2A, which encodes a ubiquitin-conjugating enzyme, is mutated in a novel X-linked mental retardation syndrome. Am. J. Hum. Genet. 79, 549–555 (2006).
Basel-Vanagaite, L. et al. Deficiency for the ubiquitin ligase UBE3B in a blepharophimosis-ptosis-intellectual-disability syndrome. Am. J. Hum. Genet. 91, 998–1010 (2012).
Frints, S. G. M. et al. Pathogenic variants in E3 ubiquitin ligase RLIM/RNF12 lead to a syndromic X-linked intellectual disability and behavior disorder. Mol. Psychiatry 24, 1748–1768 (2019).
Sobreira, N., Schiettecatte, F., Valle, D. & Hamosh, A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 36, 928–930 (2015).
Schmittgen, T. D. & Livak, K. J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108 (2008).
Labun, K., Montague, T. G., Krause, M., Torres Cleuren, Y. N., Tjeldnes, H. & Valen, E. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 47, W171–W174 (2019).
Richardson, C. D., Ray, G. J., DeWitt, M. A., Curie, G. L. & Corn, J. E. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 34, 339–344 (2016).
Modzelewski, A. J., Chen, S., Willis, B. J., Lloyd, K. C. K., Wood, J. A. & He, L. Efficient mouse genome engineering by CRISPR-EZ technology. Nat. Protoc. 13, 1253–1274 (2018).
Tesson, L., Heslan, J. M., Ménoret, S. & Anegon, I. Rapid and accurate determination of zygosity in transgenic animals by real-time quantitative PCR. Transgenic Res. 11, 43–48 (2002).
Böhm, S., Lamberti, G., Fernández-Sáiz, V., Stapf, C. & Buchberger, A. Cellular functions of Ufd2 and Ufd3 in proteasomal protein degradation depend on Cdc48 binding. Mol. Cell. Biol. 31, 1528–1539 (2011).
Baranes-Bachar, K. et al. The ubiquitin E3/E4 ligase UBE4A adjusts protein ubiquitylation and accumulation at sites of DNA damage, facilitating double-strand break repair. Mol. Cell. 69, 866–878 (2018).
Carén, H., Holmstrand, A., Sjöberg, R. M. & Martinsson, T. The two human homologues of yeast UFD2 ubiquitination factor, UBE4A and UBE4B, are located in common neuroblastoma deletion regions and are subject to mutations in tumours. Eur. J. Cancer 42, 381–387 (2006).
Zhang, M. P., Zhang, W. S., Tan, J., Zhao, M. H., Lian, L. J. & Cai, J. Poly r(C) binding protein (PCBP) 1 expression is regulated by the E3 ligase UBE4A in thyroid carcinoma. Biosci. Rep. 37, BSR20170114 (2017).
Naudin, C. et al. SLAP displays tumour suppressor functions in colorectal cancer via destabilization of the SRC substrate EPHA2. Nat. Commun. 5, 3159 (2014).
Acknowledgements
We thank the patients and families for their support and participation. This study was supported by a grant from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) (CEPID number 2013/08028-1, 2014/15982-6) and by a grant from the National Institutes of Health (NIH) Office of the Director (U42OD012210). U.S.M. was fellow of FAPESP (2016/14517-3). We also thank Euna Koo for the image in Figure S3.
Author information
Authors and Affiliations
Contributions
Conceptualization: U.S.M., D.B., F.K., H.M.B. Funding acquisition: U.S.M., M.Z., F.K., H.M.B. Methodology: U.S.M., D.B., J.A.B., K.D., J.P.K., F.K., H.M.B. Project administration: U.S.M., F.K., H.M.B. Resources: K.C.K.L., M.Z., C.F.M.M., E.S., N.D.D., F.S.A., F.K., H.M.B. Validation: B.W., L.B., B.C.L., D.J.M., F.G., F.d.S.L., D.O., F.P.M., M.T.W., A.J., M.H., H.S.A., A.C. Visualization: U.S.M., D.B., A.M., L.L. Writing—original draft: U.S.M, D.B., F.K., H.M.B. Writing—review & editing: U.S.M, D.B., F.K., H.M.B.
Corresponding authors
Ethics declarations
Ethics declaration
Studies were independently approved by the review boards of the participating institutions and performed according to local laws: family A: University of Sao Paulo CAAE 77680117.0.0000.5464; family B: Stanford University, IRB 28362; family C: King Faisal Specialist Hospital and Research Centre (KFSHRC) RAC 2121053; family D: German Gene Diagnostic Act (Gendiagnostikgesetz), EK 273072018. Written informed consent was obtained from parents or legal guardians to participate in these studies, giving permission to use patients’ (1) DNA samples for genomic sequencing, (2) blood or skin fibroblasts for establishing cell lines, and (3) photos for research publications and presentations. Mouse Model: All animal use was conducted in accordance with the Animal Welfare Act and the 2013 American Veterinary Medical Association (AVMA) Guidelines on Euthanasia. All studies were done consistent with the Institute for Laboratory Animal Research (ILAR) 8th Revision to the Guide for the Care and Use of Laboratory Animals and in compliance with and with prior approval from the University of California–Davis institutional animal care and use committee (IACUC).
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Melo, U.S., Bonner, D., Kent Lloyd, K.C. et al. Biallelic UBE4A loss-of-function variants cause intellectual disability and global developmental delay. Genet Med 23, 661–668 (2021). https://doi.org/10.1038/s41436-020-01047-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-020-01047-z