Abstract
Purpose
Most classical aniridia is caused by PAX6 haploinsufficiency. PAX6 missense variants can be hypomorphic or mimic haploinsufficiency. We hypothesized that missense variants also cause previously undescribed disease by altering the affinity and/or specificity of PAX6 genomic interactions.
Methods
We screened PAX6 in 372 individuals with bilateral microphthalmia, anophthalmia, or coloboma (MAC) from the Medical Research Council Human Genetics Unit eye malformation cohort (HGUeye) and reviewed data from the Deciphering Developmental Disorders study. We performed cluster analysis on PAX6-associated ocular phenotypes by variant type and molecular modeling of the structural impact of 86 different PAX6 causative missense variants.
Results
Eight different PAX6 missense variants were identified in 17 individuals (15 families) with MAC, accounting for 4% (15/372) of our cohort. Seven altered the paired domain (p.[Arg26Gln]x1, p.[Gly36Val]x1, p.[Arg38Trp]x2, p.[Arg38Gln]x1, p.[Gly51Arg]x2, p.[Ser54Arg]x2, p.[Asn124Lys]x5) and one the homeodomain (p.[Asn260Tyr]x1). p.Ser54Arg and p.Asn124Lys were exclusively associated with severe bilateral microphthalmia. MAC-associated variants were predicted to alter but not ablate DNA interaction, consistent with the electrophoretic mobility shifts observed using mutant paired domains with well-characterized PAX6-binding sites. We found no strong evidence for novel PAX6-associated extraocular disease.
Conclusion
Altering the affinity and specificity of PAX6-binding genome-wide provides a plausible mechanism for the worse-than-null effects of MAC-associated missense variants.
Similar content being viewed by others
INTRODUCTION
Classical aniridia (MIM 106210) is typified by absence or severe hypoplasia of the iris associated with cataracts, keratopathy, glaucoma, foveal hypoplasia, and optic nerve malformations, and absence or hypoplasia of the anterior commissure, olfactory bulb, and pineal gland.1 It is almost exclusively caused by genomic variants that result in haploinsufficiency of PAX6, by gene deletion, cis-regulatory variant, or likely/presumed gene disruptive (LGD) variants. PAX6 missense variants may mimic haploinsufficiency but are more commonly associated with milder phenotypes, such as partial aniridia, ectopia pupillae (MIM 129750), isolated foveal hypoplasia (MIM 136520), Peters anomaly (MIM 604229), or optic disc malformations.2,3,4 Compound heterozygous LGD variants of PAX6 are a rare cause of syndromic bilateral anophthalmia (absent eyes) associated with severe brain and craniofacial defects.5,6
PAX6 (MIM 607108) encodes a transcription factor essential for the correct development of the eye, central nervous system, and pancreas, and for maintenance of these tissues in adulthood.7,8,9,10 PAX6 binds DNA via two highly conserved DNA-binding domains: a homeodomain (HD) and a structurally bipartite paired domain (PD) with N-terminal (NTS) and C-terminal (CTS) subdomains, each containing three α-helices that incorporate a helix–turn–helix motif. Each subdomain can independently or cooperatively bind DNA, and can modulate the DNA-binding properties of the HD.11
The diverse range of sequences that mediate binding of PAX6 to the genome have been defined using chromatin immunoprecipitation (ChIP)12 and, subsequently, hidden Markov modeling using experimentally derived binding sites with evolutionary conservation of the bound sequences to enrich for truly functional elements.13 These approaches have identified between 600 (ref. 13) and ~2500 (ref. 12) high-confidence functional binding sites controlling between 90 (ref. 12) and 200 (ref. 12,13) different genes. The quaternary structure of the interaction of the PAX6 paired domain with a specific binding site has been determined using X-ray crystallography.14 PAX6 has been shown to bind cooperatively with SOX2 in vitro.15 Heterozygous LGD variants in SOX2 are the most common cause of severe bilateral eye malformations.16
Here we use an unbiased approach to define the phenotypic spectrum associated with PAX6 missense variants and determine how confidently the observed phenotype can be predicted by the nature and position of each causative amino acid substitution and their predicted impact on DNA binding. To define the ocular phenotypic spectrum we reviewed all of the published cases of PAX6 missense variants and those identified in the HGUeye cohort, which is enriched for severe eye malformations, particularly microphthalmia, anophthalmia, and coloboma (MAC). To look for previously undescribed extraocular phenotypes we reviewed all of the likely causative PAX6 missense variants reported in the Deciphering Developmental Disorders (DDD) study,17,18 which has performed exome sequencing on 6993 individuals with previously undiagnosed, diverse, and severe developmental disorders with adequate phenotypic data.
MATERIALS AND METHODS
Ethical approval
This study complied with the tenets of the Declaration of Helsinki: informed consent was obtained from all of the families, including permission to publish patient photos. The Medical Research Council (MRC) Human Genetics Unit eye malformation (HGUeye) cohort was collected and maintained using protocols approved by the Scotland UK Multicentre Research Ethics Committee: 06/MRE00/76 “Linkage and mutation analysis of developmental eye genes in cases of severe structural eye malformations” and 16/SS/0201 “An investigation of the genetic causes of human eye malformations.” The Deciphering Developmental Disorders (DDD) Study was collected under a protocol approved by Cambridge UK Multicentre Research Ethics Committee: 10/H0305/83 “The Deciphering Developmental Disorders Study.”
Variant analysis
Missense variants from the HGUeye cohort were identified by a variety of different methods used to perform variant analysis of the PAX6 coding sequence from genomic DNA, including heteroduplex analyses, Sanger sequencing, and targeted next-generation sequencing (including exome sequencing), essentially as described.19 PAX6 missense variants in the DDD study (www.ddduk.org), identified from trio based exome sequencing and diagnostic filtering of the variants, was performed as described.17,18 Targeted sequencing of individuals with foveal hypoplasia was performed in the Salisbury NHS Foundation Trust genetic testing program. We designated PAX6 missense variants present in the gnomAD database v.2.1 (https://gnomad.broadinstitute.org/gene/ENSG00000007372/transcript/ENST00000379132) as putatively benign for the purposes of comparison with the pathogenic variants. All variants were numbered according to the reference sequences NC_000011.10 (genomic, GRCh38), NM_000280.4; ENST00000379132 (transcript, 1269 nucleotides open reading frame), and NP_000271.1; P26367-1; LRG_720p1; ENSP00000492315.1 (isoform, 422 amino acids).
Phenotypic cluster analysis
Nonredundant components of the ocular phenotype were systematically coded in individuals with PAX6 missense variants and compared with those from a size-matched random sample of individuals with PAX6 LGD variants (Table S1). The phenotypic data were curated from original publications, and/or from the original clinical descriptions from the referring clinicians. The clustering analyses and generation of the heat maps were performed using the R packages cluster and gplots, and the function heatmap.2 (see Table S1 for further details of the phenotyping and scripting). Within heatmap.2, the functions dist and daisy were combined to compute the distance/dissimilarity matrix, respectively (combining the two allowed missing data, NA, to be taken into account in the clustering). Euclidean distance (root sum-of-squares of differences) was selected as the metric argument within daisy. When Manhattan distances (sum of absolute differences) were selected as an argument, the heat map did not change significantly (Fig. S1a).
Computational modeling and phenotype predictions
All structural analyses used the X-ray crystal structure of the DNA-bound PD (PDB ID: 6pax) and a homology model of the DNA-bound HD, built with SWISS-MODEL20 using the crystal structure of the closely related Drosophila Aristaless HD (PDB ID: 3lnq) as a template. The effects of all missense variants were modeled with the program FoldX,21 both using the protein monomer by itself and the protein:DNA complex. Default FoldX parameters were used, with ten replicates performed per variant. Variants with ΔΔG > 1.6 kcal/mol (using the same threshold described previously22) for the protein monomer were classified as “destabilize folding.” Variants with monomer ΔΔG ≤ 1.6 kcal/mol, where the difference between the ΔΔG for the full complex and the monomer was >0.25 kcal/mol, were classified as “perturb protein:DNA interaction.” All other variants were classified as “unknown molecular effect.”
In total, 21 different computational phenotype predictors were run for all of the pathogenic and putatively benign (gnomAD) variants in the primary PAX6 isoform (Uniprot ID: P26367). The predicted properties of all variants are provided in Table S2.
To assess evolutionary conservation of PAX6 residues among orthologous proteins, 124 sequences from Ensembl Compara were used (https://www.ensembl.org/Homo_sapiens/Gene/Compara_Ortholog?db=core;g=ENSG00000007372;r=11:31784779-31818062).
Electrophoretic mobility shift assay (EMSA)
EMSA experiments were performed using purified PAX6 PD protein variants and two known DNA target sequences. The PAX6 PD was expressed from plasmid vector in pGEX-6P-1t and site-directed mutagenesis was used to generate clones expressing the variants p.Cys52Arg, p.Ser54Arg, p.Arg92Gln, p.Ser121Leu, and p.Asn124Lys. The proteins were expressed as GST-fusion proteins in E. coli BL21-Gold(DE3) Competent Cells (Agilent), purified with Glutathione Sepharose 4B beads and cleaved using PreScission cleavage enzyme. DNA target sequences were double-stranded Fam-labeled oligos (Integrated DNA Technologies) incorporating the PAX6 enhancer sequences LE9 or SIMO. Reaction products were resolved on native PAGE gels and densitometry performed using Image Studio Lite (LI-COR Biosciences). Quantified binding of wild-type PAX6 PD was fitted to the Hill equation (GraphPad Prism 8 software) to calculate the Kd value, which was the concentration used for comparison of the binding properties of each of the PAX6 PD protein variants. Within each experiment, the proportion bound was normalized to the wild type. One-way repeated-measures analysis of variance (ANOVA) was used, with Dunnett’s multiple comparison test to compare the protein–DNA binding of the different protein variants versus the wild-type PAX6 PD. Alpha was set at p = 0.05.
Identification of variants in PAX6 paralogs
Eight human paralogous members of the PAX family (excluding PAX6) were selected to identify all pathogenic missense variants in the PD of each protein. Variants were identified using publicly available data from Alamut Visual v2.11 (SOPHiA Genetics), including ClinVar (pathogenic, likely pathogenic categories), and from Leiden Open Variation Database (LOVD) (http://www.lovd.nl/3.0/) and literature searching. All variants reported as pathogenic and with phenotype details were collated and tabulated in text form and as a component of a multiple sequence alignment (CLUSTAL O1.2.4).
RESULTS
Defining the spectrum of ocular phenotypes associated with PAX6 missense variants
To look for novel ocular phenotypes associated with PAX6 missense variants, we selected 372 individuals with bilateral MAC from a total of 1169 unrelated screened individuals with a wide spectrum of eye malformations in the HGUeye cohort (Table S3). In this MAC group, 17 individuals from 15 families have eight different ultrarare, heterozygous PAX6 missense variants altering seven different residues (Table 1, see Supplementary Materials and Methods for detailed clinical descriptions). In six affected individuals the variant had occurred de novo. In the two multiplex families the variant segregated appropriately with the disease. In the remaining seven affected individuals we were not able to determine the inheritance of the causative allele because all of the required samples were not available. Of the seven substituted residues in the MAC group, six were within the PD (Arg26, Gly36, Arg38, Gly51, Ser54, Asn124) and one in the HD (Asn260). Recurrent substitutions were seen in 4/6 PD residues, involving either identical (p.Arg38Trp, p.Gly51Arg, p.Ser54Arg, p.Asn124Lys) or different (p.Arg38Trp/Gln) amino acids. All individuals with a p.Ser54Arg or p.Asn124Lys variant were prescreened and found to be wild type for SOX2 and OTX2 coding sequences.
To identify missense variants that either mimic haploinsufficiency or represent likely hypomorphic alleles we took two different approaches. First we selected a group of 399 unrelated individuals from the HGUeye cohort who were strongly enriched for classical aniridia and other known PAX6-associated ocular malformations. From this analysis, seven non-MAC individuals from three families have two different heterozygous missense variants, Val78Ala and Arg128His, both of which are in the PD. The clinical features of these individuals are summarized in Table 2. In addition, we reviewed and tabulated phenotypic and genetic data on PAX6 missense variants from other sources, including all published cases that were accessible to us (Tables S1, S4), resulting in a total of 161 affected individuals from 110 families with 86 different missense variants. As a “true haploinsufficiency” comparison group, we extracted phenotypic information from a randomly selected matched number of individuals with PAX6 LGD variants (Table S1).
Phenotypic cluster analysis identifies worse-than-null PAX6 missense variants
We used the detailed ocular phenotypic and genetics information from the set of individuals identified in the previous section to perform a cluster analysis to determine whether missense and LGD variants in PAX6 could be distinguished on the basis of their phenotypes. This confirmed that aniridia is highly correlated with LGD variants. MAC and anterior segment dysgenesis cluster separately but are both correlated with missense variants (Figs. 1a, S1). Ocular features that are highly correlated with aniridia, such as foveal hypoplasia, cataract, and keratopathy, are more evenly distributed between LGD and missense variants. The individuals with p.Ser54Arg and p.Asn124Lys variants were distinguishable as severe bilateral microphthalmia with or without iris defects, coloboma, congenital corneal opacification, retinal detachment, and lens defects comprising primary aphakia, reduced size, and subluxation (Fig. 1b). This constellation is reminiscent of the ocular features associated with SOX2 variants (MIM 184429).23
(a) Quantitative phenotype-driven clustering analysis. The ocular features assessed are labeled on the x-axis. Variants encoded by pathogenic PAX6 missense variants (blue; n = 161) and a size-matched cohort of likely/presumed gene disruptive (LGD) variants (black; nonsense and frameshift variants with predicted nonsense-mediated decay) were clustered according to phenotypic features using the R packages cluster and gplots, and the function heatmap.2 (red/dark red, feature present; gray, feature absent; white, data not available). The clustering reveals that LGD variants are predominantly associated with classical aniridia features, whereas missense variants have a wider phenotypic spectrum that includes distinct clusters of microphthalmia, anophthalmia, or coloboma (MAC) spectrum and anterior segment dysgenesis phenotypes. The aniridia-associated features foveal hypoplasia, cataract, and keratopathy, which can present in the absence of iris defects, are more evenly distributed across both classes of variants. Detailed versions of the phenotyping data set used for the clustering analysis and the heat map, with all of the missense and LGD variants labeled, are available (Table S1 and Fig. S1, respectively). (b) SOX2-like phenotype associated with recurrent PAX6 p.Ser54Arg and p.Asn124Lys variants. A schematic of PAX6 isoform NP_000271.1 (P26367-1, 422 amino acids) indicating the position of the paired domain (PD; red box), homeodomain (HD; green box), the alternatively spliced 14 amino acids (yellow box) that are present in the PAX6 + 5a isoform NP_001595.2 (P26367-2, 436 amino acids), and the PAX6 coding exon boundaries (white vertical bars). The PD-HD linker region and the C-terminal transregulatory region are labeled. Images i–iv, shown above the schematic, are the eye phenotypes of HGUeye cohort MAC individuals with PAX6 p.(Ser54Arg) or p.(Asn124Lys) PD variants, which are recurrent and uniquely associated with severe bilateral defects; v–vii, shown below the schematic, are the eye phenotypes of HGUeye cohort MAC individuals with PAX6 PD (NTS) variants, which are recurrent and are associated with more variable phenotypes; v(F), fundus image of patient ID 1139 with bilateral choroidal coloboma; vii’, magnetic resonance image (MRI) showing bilateral mild microphthalmia in patient ID 4091. L left eye, R right eye.
Protein structural analysis reveals that PAX6 variants that affect DNA binding are highly associated with microphthalmia
In an attempt to understand the molecular basis underlying the MAC-associated PAX6 missense variants, we investigated their protein structural context. In total, 22/86 missense variants (32/161 affected individuals) were classified as MAC-associated in the phenotypic cluster analysis. Overall, both MAC and non-MAC variants occur throughout both halves of the PD and in the HD, and there is no obvious clustering of the MAC variants in three-dimensional space (Fig. 2a; stratified further by phenotype in Fig. S2). Ser54 and Asn124, located in helices αIII of the NTS and αVI of the CTS, both make substantial contacts with DNA, burying 44.3 and 51.8 Å2 of surface area, respectively.
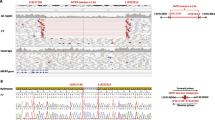
(a) Protein structural analysis of microphthalmia, anophthalmia, or coloboma (MAC) versus non-MAC missense variants. Locations of pathogenic missense variants highlighted on the structures of the DNA-binding domains: paired domain (PD) (PDB ID: 6pax) and homeodomain (HD) (homology model of the complex based upon PDB ID: 3lnq). Residues mutated in individuals with MAC are shown in red, while all other residues associated with pathogenic variants are shown in light blue. Figure S2 shows the variants further stratified by phenotype, and also shows other nonpathogenic variants from gnomAD observed in the human population. (b) Predicted molecular effects of MAC versus non-MAC missense variants. The P value is calculated with Fisher’s exact test, based upon the proportion of variants in each group of unknown molecular effect. (c) Effect of PD missense variants on binding to known DNA targets of PAX6. Electrophoretic mobility shift assays (EMSA) of the PD of PAX6. Two known DNA targets of the PAX6 PD, the LE9 and SIMO enhancer elements, were selected as binding sequences. For each element, the DNA binding of wild-type PD (lane 2 of each gel image) was compared with five different mutant PDs: Cys52Arg and Ser121Leu (C52R and S121L, aniridia-associated variants, lanes 3 and 6 of each gel image), Ser54Arg and Asn124Lys (S54R and N124K, MAC-associated variants, lanes 4 and 7 of each gel image), and Arg92Gln (R92Q, gnomAD variant, lane 5 of each gel image). For the MAC-associated variants, PD-Ser54Arg showed reduced binding to the LE9 element (left panel) but not to the SIMO element (right panel), whereas PD-Asn124Lys showed a moderate reduction in binding to both the LE9 and SIMO elements (left and right panels, respectively). Quantification data of the wild-type and mutant PD binding profiles are shown above each representative EMSA gel image: black, wild-type PD normalized binding; red, C52R and S121L aniridia-associated variants; purple, S45R and N124K MAC-associated variants; blue, R92Q gnomAD variant. (d) Comparison of disease-associated PD variants in PAX6 paralogs. A multiple sequence alignment (generated by CLUSTAL O1.2.4) showing the PD of the nine members of the PAX protein family: PAX1/9, PAX2/5/8, PAX3/7, and PAX4/6. Gray shaded blocks, α-helices αI–αIII of the N-terminal subdomain; green shaded blocks, α-helices αIV–αVI of the C-terminal subdomain; red bold, residues with pathogenic PAX6 missense variants identified in the HGUeye MAC (blue shaded), non-MAC (yellow shaded), and Deciphering Developmental Disorders (DDD) (black underlined) cohorts analyzed in this report or conserved at these positions across the protein family (unshaded); brown shaded box, residues with previously reported pathogenic missense variants (for PAX6 these are restricted to those associated with MAC phenotypes). Residues associated with MAC cases in this report are labeled below the alignment, indicating the residue number and in brackets the number of unrelated cases with a missense variant; PAX6 Ser54 and Asn124, the residues uniquely associated with severe MAC phenotypes, are shown in bold. An extended version of this figure, detailing the reported pathogenic missense variants in the HD of PAX3/7 and PAX 4/6, is available (Fig. S6). CTS C-terminal subdomain, NTS N-terminal subdomain, WT wild type.
Next, we used the molecular modeling program FoldX21 to predict the effects of PAX6 missense variants. FoldX was previously shown to be useful for identifying PAX6 variants that disrupt folding or interactions with DNA.22 We classified all 86 pathogenic variants into three categories: those predicted to be highly destabilizing to protein structure and disrupt protein folding (n = 35), those predicted to perturb the interaction with DNA (n = 22), and those of unknown molecular effect (n = 29). Interestingly, more than half (12/22) of the MAC variants were predicted to affect the interaction with DNA, compared with only 10/64 non-MAC variants (Fig. 2b). Moreover, only 1/22 MAC variants (36 individuals with MAC) had an unknown molecular effect, compared with 28/64 non-MAC variants (125 individuals with non-MAC). Other phenotypic categories showed no significant differences in their association with variants predicted to affect protein folding or DNA binding (Fig. S3, Table S2).
We also investigated the ability of many existing sequence-based phenotype predictors to identify pathogenic PAX6 missense variants (Table S5). None of them shows any significant ability to distinguish MAC from non-MAC variants. However, many perform very well at discriminating the pathogenic PAX6 missense variants from 121 putatively benign variants present in the gnomAD database. Interestingly, FoldX, which uses structure alone, is competitive with the top-ranking predictors, which rely mostly on evolutionary conservation, demonstrating the power of protein structure for predicting both the pathogenicity and phenotype of PAX6 missense variants.
EMSA characterization of variant interactions with known PAX6-binding sites
As the structural analysis suggests that many PAX6 variants are likely to affect the interaction of the PD with DNA, we used EMSA experiments to test a subset of variants for their binding to two well-characterized PAX6-binding sites: the LE9 and SIMO elements24,25 (Figs. 2c, S4). One of the severe MAC-associated variants, p.Ser54Arg, caused an 85% reduction in binding to LE9 but had no effect on SIMO binding, while p.Asn124Lys showed reduced binding to both (42% to LE9, 62% to SIMO). In contrast, the classical aniridia variants p.Cys52Arg26 and p.Ser121Leu27 both completely disrupted binding to SIMO and caused an 80–90% disruption of binding to LE9. We also tested p.Arg92Gln (rs769095184), which was presumed to be a neutral variant because it was present in eight individuals (East Asian, allele frequency 0.0004349) in gnomAD. It showed a LE9 and SIMO binding pattern very similar to p.Asn124Lys, which was initially surprising, but consistent with the fact that it is predicted by our structural analysis to moderately perturb the interaction with DNA. Interestingly, this variant was very recently reported, with limited segregation details, as being associated with classical aniridia in a Chinese family with four affected individuals.28
PAX6 missense variants as a cause of previously undescribed nonocular phenotypes
Finally, to determine whether PAX6 missense variants may cause previously undescribed conditions with significant extraocular effects, we examined the phenotypes of individuals in the DDD cohort.17,18 Seven individuals had six different ultrarare, heterozygous PAX6 missense variants (Table 3). Four individuals with structural eye anomalies had three different missense variants in the PD (p.[Arg26Gly]x2, p.[Arg26Gln]x1, p.[Gly64Ala]x1). The two different Arg26 variants are recurrent (see Table 1 and ref. 29). The remaining three individuals had either predominantly neurological phenotypes or multiple nonocular malformations. Two variants occur in the PD (p.Glu62Val and p.Ile117Thr) and are predicted to be mild at a structural level and not significantly perturb protein folding or the interaction with DNA. The other (p.Gln201His) is in the flexible linker between the PD and HD and is not particularly close to any known pathogenic variants. Interestingly, the three top-ranking computational phenotype predictors (FATHMM, M-CAP, and DeepSequence) are supportive of pathogenicity for the two PD variants, having values similar to other pathogenic PAX6 variants and distinct from most of the variants in gnomAD (Fig. S5). Overall, however, we found no clear evidence for major nonocular developmental defects due to PAX6 missense variants.
DISCUSSION
The extraordinary, almost one-to-one, association of typical aniridia to heterozygous loss-of-function PAX6 variants understandably dominates the human disease literature associated with this gene. Over many years, the MRC Human Genetics Unit has established a large research cohort of individuals with aniridia and other severe eye malformations that have been screened for causative variants in PAX6. This, together with the availability of phenotypic and trio exome sequencing data from the DDD study, has allowed us to use multiple independent approaches to define the phenotypic spectrum associated with missense variants in PAX6. This has identified a specific subset of heterozygous missense variants in the PAX6 DNA-binding domains that are associated with severe bilateral microphthalmia, most notably the recurrent substitutions of Ser54 and Asn124 within the PD.
It has been inferred that the PD arose from gene duplication of a three-helix unit, with divergent evolution resulting in the NTS and CTS losing amino acid and DNA target sequence similarity but maintaining a twofold pseudosymmetry axis that permits similar folding and docking of each subdomain into the major groove of double-stranded DNA (dsDNA). In this regard, it is interesting to note that both Ser54 and Asn124 occur at approximately equivalent positions within their subdomains and both make direct contacts with the major groove, with similar relative positioning in the 5’ and 3’ regions of the DNA target binding site (Fig. 2a). Ser54 is conserved in all members of the human PAX family, and Asn124 in PAX2/5/8 and PAX4/6 (Figs. 2d, S6). In PAX6 orthologues, Ser54 is invariant, and Asn124 is conserved in all but two avian species.
The severe bilateral microphthalmia phenotypic spectrum associated with p.Ser54Arg and p.Asn124Lys is similar to that associated with SOX2 variants (MIM 184429).23 There are several reasons this is an important observation. SOX2 is a lineage-specific transcription factor, which cobinds with Pax6 to the promoter elements of many genes in presumptive lens cells.30,31 In chicken, PAX6 and SOX2 cooperatively interact at the lens-specific DC5 enhancer of delta crystallin and the N-3 enhancer of Sox2.32,33 In an optic cup progenitor cell-conditional Sox2-deficient mouse model, the severe ocular defects, including extreme microphthalmia, are ameliorated when crossed onto a Pax6-haploinsufficient background.34 These data suggest that the transcriptional activation profiles of PAX6 and SOX2 are tightly regulated and can be interdependent, and therefore that the perturbation of the transcriptional activity of either gene can cause severe ocular defects during development. It is possible that the individuals with severe bilateral MAC and a PAX6 variant also carry an as-yet-unidentified variant in a regulatory region or upstream regulator of SOX2. However, this seems unlikely, given the number and recurrent nature of the PAX6 variants identified in the HGUeye cohort. The other important factor being that both p.Ser54Arg and all but two of the p.Asn124Lys variants occurred de novo in the affected individuals (in two adult cases inheritance could not be determined). This strongly supports monoallelic variants in PAX6 as the cause of the severe bilateral microphthalmia.
One individual, with a phenotype similar to that associated with the recurrent p.Ser54Arg and p.Asn124Lys changes, has a variant in the HD (p.Asn260Tyr). Asn260 is invariant in all paired-type HDs and is adjacent to the C-terminal basic cluster (residues 261–267) component of the PAX6 HD nuclear localization signal (Fig. S6). In zebrafish Pax6 this asparagine is critical for binding DNA target sequences and for CTS–HD interaction.35 The two previously reported MAC-associated HD variants similarly substitute conserved residues in the recognition helix, Val256 and Phe258, and cause iris defects and severe bilateral phenotypes including microphthalmia with primary aphakia or chorioretinal and optic nerve coloboma.3,36 A recently reported case with p.Arg261Gly had congenital cataract and a family history of coloboma.37 HD recognition helix variants are reported for PAX3 in association with Waardenburg syndrome type 1, and for PAX7 in a single case of cleft lip and palate (Fig. S6).38,39
Taken together, the results of the protein structural analysis and EMSA experiments have allowed us to develop hypotheses regarding the worse-than-null phenotypes we observed. The most severe MAC-associated variants perturb the interaction of any one of the three DNA-binding (sub)domains. We infer that DNA binding is unlikely to be completely abrogated as the two remaining wild-type (sub)domains are sufficient for DNA interaction. However, both the affinity of these interactions and the location of bound sites are likely to be altered. The remarkable sequence diversity of in vivo PAX6-binding sites12,13 would predict variant-specific differential effects on both the degree and repertoire of target gene activation. In contrast, the classical aniridia variants tested experimentally retain almost no DNA-binding activity, consistent with an altered dosage haploinsufficiency phenotype similar to the LGD variants. Importantly, however, these molecular properties alone cannot explain all of the phenotypic heterogeneity observed among PAX6 missense variants. This is highlighted by the fact that there are several examples in our data set of different patients with the same variant exhibiting very different phenotypes (Table S4).
Four other members of the PAX gene family are associated with robust pathogenic missense variants in the PD (Fig. 2d; see Fig. S6, Table S6 for further details). These include PAX2 (papillorenal syndrome and focal segmental glomerulosclerosis 7; MIM 120330 and 616002), PAX3 (Waardenburg syndrome type I and type 3 and craniofacial–deafness–hand syndrome; MIM 193500, 148820, and 122880), PAX8 (congenital nongoitrous hypothyroidism; MIM 218700) and PAX9 (tooth agenesis; MIM 604625). Of the eight residues that have causative substitutions detailed above in HGUeye MAC or DDD, seven are invariant across the paralogs. Asn124 is conserved only in PAX2, PAX5, and PAX8 (group II) and PAX4 (group IV); no disease-associated missense variants have been reported at this position. Six of the seven invariant residues have causative missense variants reported in PAX6 paralogs. For Ser54 a different substitution at the equivalent residue in PAX3 (p.Ser84Phe) has been reported in a single family (Table S6). Heterozygosity for this variant was not associated with a worse-than-null phenotype. Overall, of the 44 different PAX6 PD positions with pathogenic variants, 25 have pathogenic variants at equivalent positions in paralogous PAX genes. Furthermore, there are 24 PD residues with pathogenic missense variants in paralogs with no known pathogenic variants in PAX6; there is a high probability that these could be the sites of pathogenic PAX6 variants to be discovered in the future.
References
Hingorani M, Hanson I, van Heyningen V. Aniridia. Eur J Hum Genet. 2012;20:1011–1017.
Hanson I, Churchill A, Love J, et al. Missense mutations in the most ancient residues of the PAX6 paired domain underlie a spectrum of human congenital eye malformations. Hum Mol Genet. 1999;8:165–172.
Azuma N, Yamaguchi Y, Handa H, et al. Mutations of the PAX6 gene detected in patients with a variety of optic-nerve malformations. Am J Hum Genet. 2003;72:1565–1570.
Yahalom C, Blumenfeld A, Hendler K.et al. Mild aniridia phenotype: an under-recognized diagnosis of a severe inherited ocular disease. Graefes Arch Clin Exp Ophthalmol. 2018;256:2157–2164.
Glaser T, Jepeal L, Edwards JG, Young SR, Favor J, Maas RL. PAX6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous system defects. Nat Genet. 1994;7:463–471.
Schmidt-Sidor B, Szymanska K, Williamson K, et al. Malformations of the brain in two fetuses with a compound heterozygosity for two PAX6 mutations. Folia Neuropathol. 2009;47:372–382.
Yogarajah M, Matarin M, Vollmar C, et al. PAX6, brain structure and function in human adults: advanced MRI in aniridia. Ann Clin Transl Neurol. 2016;3:314–330.
Osumi N, Shinohara H, Numayama-Tsuruta K, Maekawa M. Concise review: Pax6 transcription factor contributes to both embryonic and adult neurogenesis as a multifunctional regulator. Stem Cells. 2008;26:1663–1672.
Hamasaki A, Yamada Y, Kurose T, et al. Adult pancreatic islets require differential pax6 gene dosage. Biochem Biophys Res Commun. 2007;353:40–46.
Shaham O, Menuchin Y, Farhy C, Ashery-Padan R. Pax6: a multi-level regulator of ocular development. Prog Retin Eye Res. 2012;31:351–376.
Singh S, Stellrecht CM, Tang HK, Saunders GF. Modulation of PAX6 homeodomain function by the paired domain. J Biol Chem. 2000;275:17306–17313.
Sun J, Rockowitz S, Xie Q, Ashery-Padan R, Zheng D, Cvekl A. Identification of in vivo DNA-binding mechanisms of Pax6 and reconstruction of Pax6-dependent gene regulatory networks during forebrain and lens development. Nucleic Acids Res. 2015;43:6827–6846.
Coutinho P, Pavlou S, Bhatia S, Chalmers KJ, Kleinjan DA, van Heyningen V. Discovery and assessment of conserved Pax6 target genes and enhancers. Genome Res. 2011;21:1349–1359.
Xu HE, Rould MA, Xu W, Epstein JA, Maas RL, Pabo CO. Crystal structure of the human Pax6 paired domain-DNA complex reveals specific roles for the linker region and carboxy-terminal subdomain in DNA binding. Genes Dev. 1999;13:1263–1275.
Narasimhan K, Pillay S, Huang YH, et al. DNA-mediated cooperativity facilitates the co-selection of cryptic enhancer sequences by SOX2 and PAX6 transcription factors. Nucleic Acids Res. 2015;43:1513–1528.
Williamson KA, FitzPatrick DR. The genetic architecture of microphthalmia, anophthalmia and coloboma. Eur J Med Genet. 2014;57:369–380.
Deciphering Developmental Disorders Study. Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519:223–228.
Deciphering Developmental Disorders Study. Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438.
Williamson KA, Rainger J, Floyd JA, et al. Heterozygous loss-of-function mutations in YAP1 cause both isolated and syndromic optic fissure closure defects. Am J Hum Genet. 2014;94:295–302.
Waterhouse A, Bertoni M, Bienert S, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303.
Guerois R, Nielsen JE, Serrano L. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J Mol Biol. 2002;320:369–387.
Alibes A, Nadra AD, De Masi F, Bulyk ML, Serrano L, Stricher F. Using protein design algorithms to understand the molecular basis of disease caused by protein-DNA interactions: the Pax6 example. Nucleic Acids Res. 2010;38:7422–7431.
Ragge NK, Lorenz B, Schneider A, et al. SOX2 anophthalmia syndrome. Am J Med Genet A. 2005;135:1–7.
Aota S, Nakajima N, Sakamoto R, Watanabe S, Ibaraki N, Okazaki K. Pax6 autoregulation mediated by direct interaction of Pax6 protein with the head surface ectoderm-specific enhancer of the mouse Pax6 gene. Dev Biol. 2003;257:1–13.
Bhatia S, Bengani H, Fish M, et al. Disruption of autoregulatory feedback by a mutation in a remote, ultraconserved PAX6 enhancer causes aniridia. Am J Hum Genet. 2013;93:1126–1134.
Chao LY, Mishra R, Strong LC, Saunders GF. Missense mutations in the DNA-binding region and termination codon in PAX6. Hum Mutat. 2003;21:138–145.
Chen P, Zang X, Sun D, et al. Mutation analysis of paired box 6 gene in inherited aniridia in northern China. Mol Vis. 2013;19:1169–1177.
Xiao Y, Liu X, Yang C, et al. A novel PAX6 heterozygous mutation found in a Chinese family with congenital aniridia. Genet Test Mol Biomarkers. 2019;23:495–500.
Hanson IM, Fletcher JM, Jordan T, et al. Mutations at the PAX6 locus are found in heterogeneous anterior segment malformations including Peters’ anomaly. Nat Genet. 1994;6:168–173.
Thakurela S, Tiwari N, Schick S, et al. Mapping gene regulatory circuitry of Pax6 during neurogenesis. Cell Discov. 2016;2:15045.
Hou L, Srivastava Y, Jauch R. Molecular basis for the genome engagement by Sox proteins. Semin Cell Dev Biol. 2017;63:2–12.
Saha A, Kizaki S, De D, Endo M, Kim KK, Sugiyama H. Examining cooperative binding of Sox2 on DC5 regulatory element upon complex formation with Pax6 through excess electron transfer assay. Nucleic Acids Res. 2016;44:e125.
Inoue M, Kamachi Y, Matsunami H, Imada K, Uchikawa M, Kondoh H. PAX6 and SOX2-dependent regulation of the Sox2 enhancer N-3 involved in embryonic visual system development. Genes Cells. 2007;12:1049–1061.
Matsushima D, Heavner W, Pevny LH. Combinatorial regulation of optic cup progenitor cell fate by SOX2 and PAX6. Development. 2011;138:443–454.
Bruun JA, Thomassen EI, Kristiansen K, et al. The third helix of the homeodomain of paired class homeodomain proteins acts as a recognition helix both for DNA and protein interactions. Nucleic Acids Res. 2005;33:2661–2675.
Deml B, Reis LM, Lemyre E, Clark RD, Kariminejad A, Semina EV. Novel mutations in PAX6, OTX2 and NDP in anophthalmia, microphthalmia and coloboma. Eur J Hum Genet. 2016;24:535–541.
Chauhan BK, Medsinge A, Baumgartner MP, et al. Case series: pyramidal cataracts, intact irides and nystagmus from three novel PAX6 mutations. Am J Ophthalmol Case Rep. 2018;10:172–179.
Pingault V, Ente D, Dastot-Le Moal F, Goossens M, Marlin S, Bondurand N. Review and update of mutations causing Waardenburg syndrome. Hum Mutat. 2010;31:391–406.
Leslie EJ, Taub MA, Liu H, et al. Identification of functional variants for cleft lip with or without cleft palate in or near PAX7, FGFR2, and NOG by targeted sequencing of GWAS loci. Am J Hum Genet. 2015;96:397–411.
Henderson RA, Williamson K, Cumming S, et al. Inherited PAX6, NF1 and OTX2 mutations in a child with microphthalmia and aniridia. Eur J Hum Genet. 2007;15:898–901.
Chen J, Zhu J. Analysis of PAX6 gene mutations in a Chinese family affected with congenital aniridia. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2016;33:523–525. [Article in Chinese].
Acknowledgements
We thank the affected individuals and their families who contributed to this study, Alison Brown for technical assistance with the HGUeye cohort (MRC HGU, Edinburgh), and Daniel Osborne for clinical data collection (Southampton Eye Unit). Dirk-Jan Kleinjan (University of Edinburgh) is thanked for helpful discussions and critical reading of the manuscript. K.A.W. and D.R.F. are supported by a MRC University Unit Award to the University of Edinburgh for the MRC Human Genetics Unit, H.N.H. is supported by a Wellcome Trust Training Fellowship (205171/Z/16/Z), B.J.L. is supported by the MRC Precision Medicine Doctoral Training Program, J.A.M. is supported by a MRC Career Development Award (MR/M02122X/1) and is a Lister Institute Research Prize Fellow, E.S.T. is supported by MRC and Chief Scientist Office of the Scottish Government Health Directorates Scottish Genomes Partnership funding (SGP/1) and by MRC and Engineering and Physical Sciences Research Council Molecular Pathology Node funding. The DDD study presents independent research commissioned by the Health Innovation Challenge Fund (grant number HICF-1009-003) and makes use of DECIPHER (http://decipher.sanger.ac.uk), which is funded by the Wellcome Trust (see Nature PMID: 25533962 or www.ddduk.org/access.html for full acknowledgement).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure
The authors declare no conflicts of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. If you remix, transform, or build upon this article or a part thereof, you must distribute your contributions under the same license as the original. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/.
About this article
Cite this article
Williamson, K.A., Hall, H.N., Owen, L.J. et al. Recurrent heterozygous PAX6 missense variants cause severe bilateral microphthalmia via predictable effects on DNA–protein interaction. Genet Med 22, 598–609 (2020). https://doi.org/10.1038/s41436-019-0685-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-019-0685-9
Keywords
This article is cited by
-
A novel PAX6 variant as the cause of aniridia in a Chinese patient with SRRRD
BMC Medical Genomics (2023)
-
A novel missense variant expands the phenotype and genotype of PAX6-associated foveal hypoplasia accompanied by various manifestations of anterior segment dysgenesis
BMC Ophthalmology (2023)
-
Whole-genome sequencing of multiple related individuals with type 2 diabetes reveals an atypical likely pathogenic mutation in the PAX6 gene
European Journal of Human Genetics (2023)
-
Loss-of-function, gain-of-function and dominant-negative mutations have profoundly different effects on protein structure
Nature Communications (2022)
-
PAX6 missense variants in two families with isolated foveal hypoplasia and nystagmus: evidence of paternal postzygotic mosaicism
European Journal of Human Genetics (2021)