Abstract
Purpose
To characterize clinically measurable endophenotypes, implicating the TBX6 compound inheritance model.
Methods
Patients with congenital scoliosis (CS) from China(N = 345, cohort 1), Japan (N = 142, cohort 2), and the United States (N = 10, cohort 3) were studied. Clinically measurable endophenotypes were compared according to the TBX6 genotypes. A mouse model for Tbx6 compound inheritance (N = 52) was investigated by micro computed tomography (micro-CT). A clinical diagnostic algorithm (TACScore) was developed to assist in clinical recognition of TBX6-associated CS (TACS).
Results
In cohort 1, TACS patients (N = 33) were significantly younger at onset than the remaining CS patients (P = 0.02), presented with one or more hemivertebrae/butterfly vertebrae (P = 4.9 × 10‒8), and exhibited vertebral malformations involving the lower part of the spine (T8–S5, P = 4.4 × 10‒3); observations were confirmed in two replication cohorts. Simple rib anomalies were prevalent in TACS patients (P = 3.1 × 10‒7), while intraspinal anomalies were uncommon (P = 7.0 × 10‒7). A clinically usable TACScore was developed with an area under the curve (AUC) of 0.9 (P = 1.6 × 10‒15). A Tbx6-/mh (mild-hypomorphic) mouse model supported that a gene dosage effect underlies the TACS phenotype.
Conclusion
TACS is a clinically distinguishable entity with consistent clinically measurable endophenotypes. The type and distribution of vertebral column abnormalities in TBX6/Tbx6 compound inheritance implicate subtle perturbations in gene dosage as a cause of spine developmental birth defects responsible for about 10% of CS.
Similar content being viewed by others
INTRODUCTION
Recent progress in elucidating the genetic contributions to disease and molecular etiology of clinical phenotypes presents opportunities to further subclassify human disease traits such as scoliosis according to their underlying genetic etiologies. Such genetic/genomic subclassification can result in a virtuous cycle from bench to bedside to better understand the biological perturbations underlying disease traits and pathophysiological bases of disease.1,2 To further increase the specificity of molecular diagnosis and the clinical application of genetic testing and clinical genomics, precise correlations between genotype and phenotype, allelic series, and the clinical consequences of combinations of biallelic variants at a locus need to be established.3,4 The recent report of a compound inheritance model in congenital scoliosis (CS) provides a genetic biomarker with which to potentially clinically subclassify scoliosis and define a specific clinical disease entity (TACS, TBX6-associated CS) that is associated with a particular combination of variant alleles—biallelic variants consisting of a loss-of-function (LoF) TBX6 lesion and a common risk hypomorphic allele in trans—as an underlying genetic etiology and further explore the hypothesized TBX6 gene dosage effect.5
Congenital scoliosis is a form of spinal curvature that can be caused by vertebral malformations potentially resulting from defects of formation, defects of segmentation, or a combination of the two.6 The prevalence of CS is approximately 0.5–1 per 1000 live births.7 As a major contributor to childhood and adolescent disability, CS affects patients’ lives and activities of daily living both physically and psychologically.8 CS can arise from developmental spine defects that result from perturbations in somitogenesis.7 Successful somitogenesis requires the spatial and temporal regulation of a complicated gene interaction network in which TBX6 plays an essential role. Our previous work elucidated that the TBX6 compound inheritance model accounts for approximately 7.9–10.6% of sporadic CS in the Chinese population.5 Similar analyses of independent Japanese and European CS cohorts, in multiple patients from different world populations and genetic backgrounds, supported the proposed TBX6 compound inheritance model.9,10
We previously showed the CS phenotype could result from compound inheritance,5 consisting of null variant alleles at the TBX6 locus, either a deletion copy-number variant (CNV) of the 16p11.2 region11,12 or LoF induced by single-nucleotide variants (SNVs) and indels in TBX6, which maps within the 16p11.2 region.13 With the delineation of the hypothesis of the compound inheritance and TBX6 gene dosage model,5 we were motivated to more precisely characterize the genotype-phenotype relationships of the specific genotypic combination of alleles at the TBX6 locus observed in CS cases with TBX6 variant alleles (i.e., the compound inheritance model) and the clinically observed endophenotypes characterizing the CS. In addition, we sought to explore the compound inheritance and gene dosage hypothesis in both humans and mice. Moreover, further investigations are required to test the potential utility of molecular diagnosis and clinical genomics in the precision medicine–directed clinical evaluation and management of CS.
MATERIALS AND METHODS
Participants
We recruited two independent cohorts and a multicenter case series from China, Japan, and the United States. The discovery set (cohort 1) consists of unrelated sporadic CS patients of Chinese Han descent in the DISCO (Deciphering disorders Involving Scoliosis and COmorbidities, http://discostudy.org/) study from Peking Union Medical College Hospital (PUMCH) between October 2010 and January 2016, in which clinical diagnoses of CS were confirmed by radiological imaging.5 The first replication cohort (cohort 2) comprised Japanese patients with CS recruited from Japan, in compliance with the selection criteria as reported previously.10 The second replication case series (cohort 3) comprised patients with 16p11.2 deletion/TBX6 LoF variants collected from the United States. Genomic DNA was extracted from the peripheral blood or saliva samples. Informed consent was obtained from each participant or the corresponding guardian. The study was approved by the institutional review boards of PUMCH, RIKEN, Baylor College of Medicine, and the other participating hospitals.
Genetic analyses
A genome-wide CNV analysis was performed in 20 sporadic CS patients in cohort 15. Quantitative polymerase chain reaction (qPCR) analysis was conducted to screen for 16p11.2 deletions in the remaining 325 patients from cohort 1 and in the 142 patients from cohort 2. An orthogonal experimental approach to measure copy-number alteration, customized comparative genomic hybridization (CGH) microarrays or digital droplet PCR (ddPCR), was used to independently confirm the deletion CNV candidates. The 16p11.2 deletion case series (cohort 3) was identified by chromosomal microarray analysis (CMA) (Table S1).12,14,15
The entire TBX6 gene and its upstream region were amplified and analyzed by Sanger sequencing in all patients from cohort 15 and 121 patients from cohort 210. TBX6 variants were also detected by exome sequencing (ES) and verified by Sanger sequencing in the remaining 21 patients from cohort 2 and 1 patient (BH8084) from cohort 3 (Table S1). Based on our previous findings,5 we analyzed the hypomorphic allele in patients with 16p11.2 deletions and TBX6 LoF variants.
Phenotype evaluation
We conducted detailed phenotypic analyses of the spine, ribs, and intraspinal anomalies in cohort 1 and cohort 2. The age of onset indicated the age at which an individual first came to medical attention, exhibiting or presenting with features such as asymmetric shoulder height or shoulder blade prominence. Morphologically, the vertebral malformations were classified as defects of formation, defects of segmentation, or mixed malformations.16 Variations in the number of vertebral bodies were also described. The incidence of rib defects, along with the rib structure and the number of alterations, were also analyzed. The rib defect subtypes were classified as simple or complex.17 Intraspinal anomalies were defined as any defect involving the spinal cord, such as a tethered cord or syringomyelia.
Generation and evaluation of the Tbx6 gene-edited mice
Zygotes from FVB mice were edited by CRISPR/Cas918 to generate a Tbx6 frameshift allele and in independent experiments a Tbx6 hypomorphic allele.19 Matings of strains with different Tbx6 genotypes were performed to derive the Tbx6 locus biallelic variant mouse model with a Tbx6 LoF variant in exon 2 in trans with the hypomorphic haplotype in the T binding site in the promoter of Tbx6 on the second allele (Tbx6wt/- X Tbx6wt/mild-hypomorphic(mh) to derive Tbx6-/mh).19 Multiple adult animals (N = 92, 35–45 days) were evaluated by micro computed tomography (micro-CT) (Bruker, Belgium) to assess the phenotypes; this included 52 with compound inheritance, Tbx6-/mh. No randomization was used and no blinding was done in the animal study.
Development of the TACS predictive model
We developed a multivariate model of the risk score (TACScore) to clinically predict TACS. The TACScore was derived from cohort 1 (training data set) by binary logistic regression,20 in which all reliable variables that were significantly associated (P < 0.05) with TACS were entered into a multivariate model to discover the optimal predictors for distinguishing TACS from all the enrolled subjects with CS. A scoring system was developed for making these complex statistical models useful to clinicians and to evaluate the clinical utility of the TACScore, in which the point of each predictor was assigned according to the product of the corresponding β coefficient and value of the predictors with point totals corresponding to the risk estimate.20 The receiver operating characteristic (ROC) was used to assess the effectiveness. Cutoff points for the TACScore were determined with the Youden index [J=maxc(sensitivity+specificity–1)]21 to define the maximum potential effectiveness of the predictive models. Then the TACScore was validated independently in cohort 2 (testing data set).
Statistical analysis
The Mann–Whitney U test was used to analyze the ages, and the numbers of vertebral and rib malformations. The gender association and prevalence of different types of vertebral malformations were compared using Pearson's χ2 test or Fisher's exact test. The odds ratio (OR) with 95% confidence interval (CI) was used to assess the influences of TBX6 LoF variants on vertebral, rib, and intraspinal malformations. Statistical analyses were performed with SPSS version 15.0 (SPSS, USA). A value of P < 0.05 was considered statistically significant. The variance was similar between the groups that are statistically compared. (Methodological details are provided in the Supplementary Materials.)
RESULTS
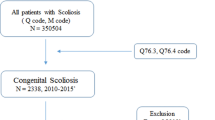
We enrolled 345 Chinese patients with sporadic CS as cohort 1 (156 males and 189 females, 12 years [7–15] as the median age at enrollment [the interquartile range, IQR], 3 years [1–9] at onset, Figure S1), including 237 previously reported5 and 108 new CS patients (Fig. 1). For cohort 2, 142 Japanese patients with CS were recruited in Japan (79 previously reported10 and 63 new patients, Fig. 1). Ten patients with evidence for compound inheritance, 16p11.2 deletion/TBX6 LoF variants, who were systematically evaluated for vertebral phenotypes were enrolled in cohort 3 (Fig. 1 and Table 1). Collectively, 26 patients with proximal 16p11.2 deletions, 5 patients with frameshift variants (c.1250_1251insT, c.266_267insC, c.704_705insG, c.1169_1170insC, and c.1179_1180delAG), and 2 patients with stop-gain variants (c.844C>T [p.R282*], c.933C>A [p.C311*]) in the TBX6 gene were identified in cohort 1 (Table 1, Fig. 1, Figure S2–4, and Table S2). Similarly, ten 16p11.2/TBX6 deletions, two TBX6 frameshift variants (c.156delG and c.935_936insGA), one nonsense variant (c.699G>A [p.W233*]), one splice-site variant (c.119-1G>A), and one novel missense variant (c.333G>T [p.M111I]), which had been confirmed as a LoF variant,10 were identified in cohort 2 (Fig. 1, Table 1, Figure S4B, and Table S2). Further haplotype analysis showed that all patients with TBX6 LoF variants had the T-C-A risk haplotype (the co-occurrence of three common single-nucleotide polymorphisms [SNPs], namely, rs2289292, rs3809624, and rs38096275,22) in trans on the opposite allele in cohort 1 and cohort 2 (Table 1). Thus, the percentage of new CS patients explained by the TBX6 compound inheritance model (10/108 [9.3%] in cohort 1 and 15/142 [10.6%] in cohort 2) was consistent with our previous findings (23/237 [9.7%])5. In cohort 3, ten patients with TBX6 deleterious variants (nine 16p11.2 deletions and one frameshift variant [c.469_470insCGGC, p.R157fs], Figure S4C, Table 1) were identified. Furthermore, we identified 13 patients with 14 TBX6 missense variants and 1 patient with an in-frame insertion variant from cohort 1; these 14 subjects were excluded from the in-depth systematic phenotypic and clinical endophenotypic data analyses described below as the functional effects of these variants were uncertain.
Workflow for TBX6-associated congenital scoliosis (TACS) study participants in the multicenter cohorts and the animal model genotype-phenotype analyses. The discovery set (cohort 1) consisted of 345 unrelated Chinese patients with sporadic CS. The status of the TBX6 variants was screened, and the clinical characteristics were reviewed and compared according to the TBX6 genotypes, including vertebral, rib, and intraspinal anomalies. To recapitulate and investigate the phenotypic consequences of the compound inheritance and TBX6/Tbx6 gene dosage genetic model, a mouse strain with a specific combination of Tbx6 alleles was constructed. Tbx6 alleles were individually engineered and a mouse strain for the compound inheritance model was constructed by mating, introducing a truncated allele in trans with a mild hypomorphic (mh) allele, Tbx6-/mh. Furthermore, the TBX6-associated CS score (TACScore) was developed to guide and increase the efficiency of diagnosing TACS from clinically observed endophenotypes. CS congenital scoliosis, LoF loss-of-function, Non-TACS non-TBX6-associated CS, TACS TBX6-associated CS, VUS variants of unknown significance. aCohort 1 comprised the 237 sporadic CS patients enrolled between October 2010 and June 2014 in our previous study.5 bCohort 2 comprised 79 Japanese CS patients from 94 previously described CS patients,10 after excluding 15 patients without complete clinical data. cNo information regarding whether the risk haplotype was in trans or in cis with the TBX6 frameshift variant.
Distinct endophenotype of patients with TACS
We divided patients in cohort 1 into two groups based on the TBX6 genotype. The 33 CS patients containing TBX6 LoF variants and the in trans T-C-A risk haplotype were classified as those with compound inheritance at TBX6 and their form of CS as TACS, and the remaining 298 patients were classified as non-TACS (Fig. 1).
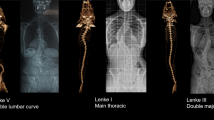
The TACS patients were significantly younger than the non-TACS at the age of onset (TACS, 2 years [1–3]; non-TACS, 3 years [1–9]; P = 0.02 by the Mann–Whitney U test; Table 2). A higher proportion of male than female was found in the TACS group in cohort 1 (TACS, 21/33 [63.6%]; non-TACS, 128/298 [43.0%]; P = 0.03 by the Pearson χ2 test; Table 2). Overall, the vertebral anomalies were less complex in the TACS group as defined by the number of vertebral malformations (TACS, 2 [1–2]; non-TACS, 4 [2–6]; P = 2.8 × 10‒9; Table 2 and Fig. 2). Remarkably, all spinal deformities in the TACS group originated from the defect of vertebral formation, in which 29 (87.9%) had a simple type (P = 9.2 × 10‒14, Table 2 and Fig. 2). Specifically, all TACS patients exhibited hemivertebrae or butterfly vertebrae (TACS, 33/33 [100%]; non-TACS, 171/298 [57.4%]; P = 4.9 × 10‒8; OR 1.7; 95% CI 1.6–1.9; Table 2 and Figure S6A). More segmented hemivertebrae/butterfly vertebrae (TACS, 1 [1–1.5]; non-TACS, 0 [0–1]; P = 1.2 × 10‒12; Table 2 and Fig. 2) and fewer block vertebrae (TACS, 0 [0–0]; non-TACS, 0 [2–4]; P = 8.8 × 10‒10; Table 2 and Fig. 2) were noted in the TACS group.
Comparison of the distribution regarding abnormal vertebrae in non-TACS and TACS patients and in Tbx6-/mh mouse compound inheritance and gene dosage model. The x-axis shows the frequency of each malformation in each vertebra and the y-axis shows the vertebral distribution in the spine. The vertebral malformations in the non-TACS group (N = 298) were normally distributed. There were 58 TACS patients in total in three worldwide cohorts (cohort 1 from Peking Union Medical College Hospital [PUMCH] in China, N = 33; cohort 2 from the multiple centers in Japan, N = 15; cohort 3 from multiple centers in the USA, N = 10). The Tbx6-/mh engineered mice (N = 52) exhibited a distinct phenotype, namely, a defect of formation involving the lower part of the spine. In addition, malformations in the upper and middle thoracic spine were significantly less involved (P = 7.3 × 10‒5 and 3.0 × 10‒6, respectively) in the TACS group than in the non-TACS group. C cervical vertebra, CS congenital scoliosis, L lumbar vertebra, mh mild hypomorphic, Non-TACS non-TBX6-associated CS, S sacral vertebra, T thoracic vertebra, TACS TBX6-associated CS.
Of note, the vertebral segment location and spine level distribution regarding the type of abnormal vertebrae in the TACS group were distinct from that of the non-TACS group (Fig. 2). In the TACS group, the lower part of the spine (T8–S5) was more frequently involved (TACS, 33/33 [100%]; non-TACS, 247/298 [82.9%]; P = 4.4 × 10‒3; Table 2 and Fig. 2). While the incidence of rib anomalies was comparable between the two groups, more simplified rib anomalies (e.g., a localized fusion of two ribs or an absence of one rib) versus complex abnormalities (e.g., multiple extensive rib fusions or adjacent large chest wall defects) were more frequent in the TACS patients (TACS, 25/33 [75.8%]; non-TACS, 88/298 [29.5%]; P = 3.1 × 10‒7; Table 2 and Figure S6B). Furthermore, fewer intraspinal malformations (e.g., tethered cords or syringomyelia) were observed in the TACS group (TACS, 1/33 [3.0%]; non-TACS, 131/298 [44.0%]; P = 7.0 × 10‒7; Table 2).
Tbx6 gene-edited mouse model for compound inheritance
We engineered the Tbx6 LoF variant (Tbx6‒) by introducing a 1-bp insertion in exon 2. The Tbx6mh allele was generated as a functional equivalent to the human mild hypomorphic allele (Figure S7)19. Gene expression in vitro was downregulated by the Tbx6mh mutant to approximately 65% of the wild-type gene, which was close to the 70% dosage level of the human TBX6 mild hypomorphic allele (Figure S7C)19. These engineered Tbx6 alleles were used to construct strains with different genotypic combinations.
As predicted, and consistent with literature observations, no homozygotes for the LoF variant were identified in liveborn animals and expected Mendelian ratios of particular genotypic combinations were distorted in liveborns,19 consistent with embryonic lethality of the Tbx6-/- null animals.23 We acquired through genetic matings and phenotypically assessed mice with five specific genotypes: Tbx6wt/wt (N = 10; wt, wild-type), Tbx6wt/- (N = 10), Tbx6wt/mh (N = 10), Tbx6mh/mh (N = 10), and Tbx6-/mh (N = 52). Consistently, only the Tbx6-/mh mice exhibited vertebral malformations (48/52 [92.3%], P = 9.3 × 10‒9 [OR, 13.0; 95% CI, 5.1–33.3], Figure S8 and Table S3). Intriguingly, as observed in human TACS patients, all Tbx6-/mh mice had vertebral malformations involving the lower part of the spine (Fig. 2 and Table S3). Defects of vertebral column formation were present in most of the Tbx6-/mh mice (36/52 [69.2%], P = 4.9 × 10‒5 [OR, 3.3; 95% CI, 2.2–4.9], Fig. 2 and S8 and Table S3). The recapitulation of the type, extent, and distribution of vertebral malformations in the engineered compound inheritance model in mice (Fig. 2) further supports the compound inheritance and gene dosage model for TACS and implicates biological perturbations in vertebral column malformations in this type of CS.
Worldwide multicenter replications
For the first replication cohort (cohort 2), we recruited 142 CS patients from Japan and identified 15 TACS patients (Fig. 1, Table 1, and Table S2). As expected, all TACS patients had one or more hemivertebrae/butterfly vertebrae (TACS in cohort 2, 15/15 [100%]; non-TACS, 99/127 [78.0%]; P = 0.04 [OR, 1.3; 95% CI, 1.2–1.4]). All TACS patients in cohort 2 exhibited malformed vertebrae at the lower part of the spine (T8–S5), which is consistent with the patterns observed in cohort 1 (Table 1, Table S2, and Fig. 2). This pattern, of vertebral malformations and distribution of defects in the vertebral column, was observed only in the mice with compound inheritance and gene dosage perturbations below that of haploinsufficiency (i.e., Tbx6wt/-), but not equivalent to the Tbx6-/- combination of alleles.
Furthermore, we collected another replication case series (cohort 3) containing ten TACS patients in the United States. Intriguingly, hemivertebrae/butterfly vertebrae involving the lower half of the spine were again observed. The replication of our observations for this distinct genotypic combination in two additional independent cohorts from distinct world populations provides substantial evidence in support of the compound inheritance and TBX6 gene dosage model and speaks to the universality of TACS and the compound inheritance model in human and medical genetics.
TACScore to clinically predict TACS
We developed a model to predict TACS from the phenotypic data, and clinically measurable endophenotypes, obtained from cohort 1 by logistic regression.20 The final multivariate risk model, the TACScore, integrated variables including (1) segmented hemivertebrae/butterfly vertebrae involving the lower half of the spine (T8–S5), (2) the number of vertebral malformations, (3) the presence of intraspinal defects, and (4) the type of rib defect (Fig. 3a and Table S4). The calculated score of each variant parameter was linearly correlated with the corresponding risk in the regression model and defined from –8 to 4 (Fig. 3b). The cutoff point was selected as ≥3 to achieve the highest Youden index (J = 84.8%), with sensitivity, specificity, and accuracy levels higher than 90% (Table S5). The area under the curve (AUC) of the ROC curve was 0.9 (P = 1.6 × 10‒15; 95% CI, 0.9–1.0; Fig. 3c). Importantly, we validated the TACScore in cohort 2 with an AUC of 0.8 (P = 1.5 × 10‒4; 95% CI, 0.7–0.9; Fig. 3c and Table S6). Therefore, we introduced a clinical assessment pipeline for CS to efficiently identify high-risk patients for TACS and potential compound inheritance at the TBX6 locus (Fig. 3d).
Development and validation of the risk prediction model and diagnostic pipeline of TACS. a TACScore: risk predictive model for TACS. The final multivariate risk model was developed through a binary logistic regression analysis of detailed phenotypic data from Cohort 1. b The predictive efficacy of the TACScore. The x-axis shows the spectrum of TACScore and y-axis shows the predicted TACS frequency and the percentage of TACS patients in all CS patients with each calculated score in cohort 1 and cohort 2. The TACScore presented excellent predictive efficacy by comparing the predicted TACS risk with the real TACS frequency. The cutoff point was selected as 3 to achieve the highest accuracy. c Receiver operating characteristic (ROC) curve for the TACScore in cohort 1 and cohort 2. Areas under the curve (AUCs) were 0.9 (P = 1.6 × 10‒15; 95% CI, 0.9–1.0) for the discovery cohort (cohort 1) and 0.8 (P = 1.5 × 10−4; 95% CI, 0.7–0.9) for the validation cohort (cohort 2). d A proposed guideline for predicting and evaluating TACS. The risk of TACS evaluated by TACScore is suggested to perform prior to genetic testing. After the detection of TACS, a systemic evaluation with early interventions and genetic consultation were recommended. CMA chromosomal microarray analysis, CS congenital scoliosis, ES exome sequencing, GS genome sequencing, TACS TBX6-associated CS. aTBX6 compound variant contains a 16p11.2 deletion/TBX6 loss-of-function variant in the compound heterozygous configuration with the risk haplotype providing a hypomorphic variant. bVUS, variants with unknown significance. cEvidence from large-scale case–control studies, pedigree analysis, and functional studies are needed.
DISCUSSION
In this study, we expanded our knowledge of the contribution of the TBX6 compound inheritance to CS by providing international patient cohort studies and animal model–based evidence that (1) 9.6% (33/345) and 10.6% (15/142) of the patients in cohort 1 and cohort 2, respectively, could be parsimoniously explained by the TBX6 compound inheritance gene dosage model (Table 1 and Fig. 1); (2) all patients in the three cohorts had the risk haplotype in trans with a TBX6 LoF variant, with the exception of one patient (BCM01) for whom haplotype information was not available and one patient (BH8084) for whom allelic phasing of the identified variants was not available (Table 1); and (3) only the Tbx6-/mh genotype presented with vertebral malformations in the gene-edited mice (Fig. 2 and S8 and Table S3). Moreover, we elucidated a novel genetically and phenotypically defined disease entity, which carries a TBX6 LoF variant in trans with the hypomorphic allele and manifests a distinguishable constellation of clinical features and CS endophenotypes. Furthermore, we developed the TACScore system and proposed a clinical practice algorithm (Fig. 3) for evaluating CS and predicting TACS. Finally, we show qualitatively the same kind (hemivertebrae/butterfly vertebrae) and quantitatively the same distribution (lower spine) of vertebral malformations with the compound inheritance combination of variant alleles and TBX6/Tbx6 gene dosage model in human and mice.
TBX6 is a member of the T-box family.24 Variants in Tbx6 lead to phenotypes of vertebral and rib defects in mice.23,25 Tbx6 is important for cell fate in the paraxial mesoderm structure26 and formation of the segmental boundary,27 which is the precursor of the vertebral column.28 Therefore, the genetic and developmental malfunction in Tbx6 perturbation of the gene dosage model may parsimoniously explain the hemivertebrae/butterfly vertebrae presenting in all TACS patients (Table 1 and Fig. 2). Interestingly, Tbx6-/- produces an embryonic lethal phenotype with expanded tailbud and lack of posterior somites,23 suggesting that Tbx6 functions to maintain the specification of the posterior paraxial mesoderm.7 In zebrafish, tbx6 is required in the posterior of the embryo during the posterior part of primary neurulation and all of the secondary neurulation.29,30 Therefore, this developmental process optimally explains the observation that all TACS patients and Tbx6-/mh gene-edited mice show vertebral malformations anatomically localized at the lower part of the spine.
TACS does not follow “conventional” rare variant Mendelian inheritance expectations likely reflecting the embryonic lethality of TBX6/Tbx6 homozygous null alleles. Instead, TACS is caused by biallelic variants at a locus, consistent with autosomal recessive trait manifestation, but the presence of a single rare LoF variant in trans with one common hypomorphic allele; a gene dosage that is less than haploinsufficiency but not zero as with homozygous null alleles. This genetic model (compound inheritance and TACS) may provide an explanation for the scenarios in which monoallelic variants and phenotypes are perceived to be following a dominant inheritance pattern but for a disease trait with incomplete penetrance or the disease pattern observed is pseudodominance as found when the carrier state occurs at a high frequency in the population. Because the T-C-A haplotype is common worldwide (44% among Asians and 33% among Europeans, but <1% among Africans31), the haplotype is likely to be parsed and filtered out by the current genomic analytical pipelines. Therefore, for some single-locus genetic models, the functional effects of individual variant alleles, as well as the combination of alleles, should be considered when interpreting the genetic data and specific phenotype observed for the “unsolved” cases. Moreover, the compound inheritance model may be particularly relevant to recurrent rearrangement CNV32 loci with relatively high mutational frequency rates33 wherein deletion CNV can result in haploinsufficiency for many gene loci (e.g., the 0.6-Mb 16p11.2 deletion CNV contains 27 annotated genes).5,15 It is important to know not only the functional effects of individual variant alleles, but also the functional consequences of combinations of variant alleles at a locus.
A significantly small proportion of the TACS patients in cohort 1 were female (Table 2, 12/33 [36.4%] in the TACS patients and 170/298 [57.0%] in the non-TACS patients in cohort 1), which might be due to the interference of the 16p11.2 deletion and TBX6 variants in the formation of the female reproductive system.34 Notably, pleiotropic effects of 16p11.2 deletion CNV include other traits related to obesity,35 cognitive phenotypes including intellectual disability and psychiatric disorders,15,36 birth defects such as CAKUT (congenital anomalies of the kidney and urinary tract),37 and may even be involved in other undiscovered phenotypes or diseases. Thus, TACS patients are recommended to be evaluated systematically with long-term follow-up (Fig. 3d).
More importantly, from an orthopedic surgeon’s clinical perspective, hemivertebrae/butterfly vertebrae are the most remarkable features of TACS (Fig. 2). Surgical intervention and treatment at an early age results in a satisfactory prognosis for the patient and ameliorates scoliosis.38 For example, patient XH101 was diagnosed as having CS with an L2 hemivertebra at 3 years of age, and molecular testing indicated that she had a frameshift variant in TBX6 with the in trans T-C-A risk haplotype. The TACS diagnosis was made by integrating the molecular testing and clinical phenotypes. We performed L2 hemivertebra resection with short segment internal fixation, and removed the instrument at 3 years postoperation when she had a balanced spine. The last follow-up, which was at 9 years postoperation, showed an almost completely normal growth of the spine and normal curvature of the spinal column (Figure S9).
To realize clinical applications from bench to bedside, we developed a TACScore prediction algorithm as an objective adjuvant clinical measure to guide clinical management for patients with CS. The TACScore was validated in two independent cohorts and further integrated into the proposed guidelines for evaluating the risk of TACS (Fig. 3). Considering the possibility of phenotypic expansion,4 the prediction algorithm should include multiple variables associated with TACS comprehensively (i.e., endophenotypes) instead of relying on one particular phenotype (e.g., scoliosis). This clinical phenotypic assessment provides efficient information to physicians and families regarding whether the patient is suitable for 16p11.2/TBX6 variant detection or a genome-wide test. Clinical ES39 or genome-wide analysis are reserved for cases with low-risk TACScores or high-risk TACScores that are negative for 16p11.2/TBX6 pathogenic variants. Furthermore, the cutoff point of the TACScore could be adjusted according to medical practice experience and continued future observations as well as the local medical resources available to achieve the maximum health economic benefits.
Conclusions
We have defined a new subtype of CS, i.e., TACS, containing a TBX6 LoF variant compound with a hypomorphic allele. The TACS is a clinical entity defined by consistent clinically measurable endophenotypes: i.e., younger age at onset, hemivertebrae/butterfly vertebrae involving the lower part of the spine, simple rib anomaly, and fewer vertebrae and intraspinal defects. The TACScore can guide clinical management and genetic and clinical genomic testing. Human and mouse studies further confirm the compound inheritance and gene dosage model, and provide insights into potential biological consequences for spine development of perturbations in TBX6/Tbx6 gene dosage and expression. Such genetic models may be important to other birth defects.
References
Manolio TA, Fowler DM, Starita LM, et al. Bedside back to bench: building bridges between basic and clinical genomic research. Cell. 2017;169:6–12.
Yang Y, Muzny DM, Xia F, et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–1879.
Maver A, Lovrecic L, Volk M, et al. Phenotype-driven gene target definition in clinical genome-wide sequencing data interpretation. Genet Med. 2016;18:1102–1110.
Karaca E, Posey JE, Coban Akdemir Z, et al. Phenotypic expansion illuminates multilocus pathogenic variation. Genet Med. 2018 Apr 26; https://doi.org/10.1038/gim.2018.33 [Epub ahead of print].
Wu N, Ming X, Xiao J, et al. TBX6 null variants and a common hypomorphic allele in congenital scoliosis. N Engl J Med. 2015;372:341–350.
Hedequist D, Emans J. Congenital scoliosis. J Am Acad Orthop Surg. 2004;12:266–275.
Sparrow DB, Chapman G, Smith AJ, et al. A mechanism for gene-environment interaction in the etiology of congenital scoliosis. Cell. 2012;149:295–306.
Shen J, Wang Z, Liu J, Xue X, Qiu G. Abnormalities associated with congenital scoliosis: a retrospective study of 226 Chinese surgical cases. Spine (Phila Pa 1976). 2013;38:814–818.
Lefebvre M, Duffourd Y, Jouan T, et al. Autosomal recessive variations of TBX6, from congenital scoliosis to spondylocostal dysostosis. Clin Genet. 2017;91:908–912.
Takeda K, Kou I, Kawakami N, et al. Compound heterozygosity for null mutations and a common hypomorphic risk haplotype in TBX6 causes congenital scoliosis. Hum Mutat. 2017;38:317–323.
Shen Y, Chen X, Wang L, et al. Intra-family phenotypic heterogeneity of 16p11.2 deletion carriers in a three-generation Chinese family. Am J Med Genet B Neuropsychiatr Genet. 2011;156:225–232.
Al-Kateb H, Khanna G, Filges I, et al. Scoliosis and vertebral anomalies: additional abnormal phenotypes associated with chromosome 16p11.2 rearrangement. Am J Med Genet A. 2014;164A:1118–1126.
Sparrow DB, McInerney-Leo A, Gucev ZS, et al. Autosomal dominant spondylocostal dysostosis is caused by mutation in TBX6. Hum Mol Genet. 2013;22:1625–1631.
Shen Y, Irons M, Miller DT, et al. Development of a focused oligonucleotide-array comparative genomic hybridization chip for clinical diagnosis of genomic imbalance. Clin Chem. 2007;53:2051–2059.
Shinawi M, Liu P, Kang SH, et al. Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. J Med Genet. 2010;47:332–341.
McMaster MJ, Ohtsuka K. The natural history of congenital scoliosis. A study of two hundred and fifty-one patients. J Bone Joint Surg Am. 1982;64:1128–1147.
Tsirikos AI, McMaster MJ. Congenital anomalies of the ribs and chest wall associated with congenital deformities of the spine. J Bone Joint Surg Am. 2005;87:2523–2536.
Platt RJ, Chen S, Zhou Y, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455.
Yang N, Wu N, Zhang L, et al. TBX6 compound inheritance leads to congenital vertebral malformations in humans and mice. Hum Mol Genet. 2018 Oct 10; https://doi:10.1093/hmg/ddy358 [Epub ahead of print].
Sullivan LM, Massaro JM, D'Agostino RB Sr. Presentation of multivariate data for clinical use: the Framingham Study risk score functions. Stat Med. 2004;23:1631–1660.
Reiser B. Measuring the effectiveness of diagnostic markers in the presence of measurement error through the use of ROC curves. Stat Med. 2000;19:2115–2129.
Liu J, Zhou Y, Liu S, et al. The coexistence of copy number variations (CNVs) and single nucleotide polymorphisms (SNPs) at a locus can result in distorted calculations of the significance in associating SNPs to disease. Hum Genet. 2018;137:553–567.
Chapman DL, Papaioannou VE. Three neural tubes in mouse embryos with mutations in the T-box gene. Tbx6 Nat. 1998;391:695–697.
Papapetrou C, Putt W, Fox M, Edwards YH. The human TBX6 gene: cloning and assignment to chromosome 16p11.2. Genomics. 1999;55:238–241.
White PH, Farkas DR, McFadden EE, Chapman DL. Defective somite patterning in mouse embryos with reduced levels of Tbx6. Development. 2003;130:1681–1690.
Chapman DL, Agulnik I, Hancock S, Silver LM, Papaioannou VE. Tbx6, a mouse T-Box gene implicated in paraxial mesoderm formation at gastrulation. Dev Biol. 1996;180:534–542.
Zhao W, Ajima R, Ninomiya Y, Saga Y. Segmental border is defined by Ripply2-mediated Tbx6 repression independent of Mesp2. Dev Biol. 2015;400:105–117.
Dahmann C, Oates AC, Brand M. Boundary formation and maintenance in tissue development. Nat Rev Genet. 2011;12:43–55.
Copp AJ, Greene ND, Murdoch JN. The genetic basis of mammalian neurulation. Nat Rev Genet. 2003;4:784–793.
Nikaido M, Kawakami A, Sawada A, Furutani-Seiki M, Takeda H, Araki K. Tbx24, encoding a T-box protein, is mutated in the zebrafish somite-segmentation mutant fused somites. Nat Genet. 2002;31:195–199.
The 1000 Genomes Project Consortium, Abecasis GR, Auton A, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65.
Lupski JR. Structural variation mutagenesis of the human genome: impact on disease and evolution. Environ Mol Mutagen. 2015;56:419–436.
Lupski JR. Genomic rearrangements and sporadic disease. Nat Genet. 2007;39 7 suppl:S43–47.
Sandbacka M, Laivuori H, Freitas E, et al. TBX6, LHX1 and copy number variations in the complex genetics of Mullerian aplasia. Orphanet J Rare Dis. 2013;8:125.
Bachmann-Gagescu R, Mefford HC, Cowan C, et al. Recurrent 200-kb deletions of 16p11.2 that include the SH2B1 gene are associated with developmental delay and obesity. Genet Med. 2010;12:641–647.
Weiss LA, Shen Y, Korn JM, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675.
Verbitsky M, Westland R, Perez A, et al. The copy number variation landscape of congenital anomalies of the kidney and urinary tract. Nat Genet. (in press).
Guo J, Zhang J, Wang S, et al. Surgical outcomes and complications of posterior hemivertebra resection in children younger than 5 years old. J Orthop Surg Res. 2016;11:48.
Yang Y, Muzny DM, Reid JG, et al. Clinical whole-exome sequencing for the diagnosis of Mendelian disorders. N Engl J Med. 2013;369:1502–1511.
Acknowledgements
We thank all the individuals, families, and physicians involved in the study for their participation. We thank the nurses from the Department of Orthopedic Surgery at Peking Union Medical College Hospital for assistance with patient enrollment. We thank the members of Japan Early Onset Scoliosis Research Group: Noriaki Kawakami, Toshiaki Kotani, Hideki Sudo, Ikuho Yonezawa, Koki Uno, Hiroshi Taneichi, Kei Watanabe, Shohei Minami, Hideki Shigematsu, Ryo Sugawara, Yuki Taniguchi, and Nao Ootomo. We thank Sally Dunwoodie and Gavin Chapman from Victor Chang Cardiac Research Institute for their collaborations. We appreciate the support of Ellen Wald at the University of Wisconsin–Madison. This research was funded in part by the National Natural Science Foundation of China (81822030 and 81501852 to N.W., 81472045 and 81772301 to G.Q., 31625015, 31571297 and 31771396 to F.Z., 81472046 and 81772299 to Z.W., 31521003 to L.J., 81672123 to J.Z., 7162029 to X.C.), Beijing Natural Science Foundation (7172175 to N.W., 7162029 to X.C.), Beijing Nova Program (Z161100004916123 to N.W.), Beijing Nova Program Interdisciplinary Collaborative Project (xxjc201717 to N.W.), 2016 Milstein Medical Asian American Partnership Foundation Fellowship Award in Translational Medicine (to N.W.), the Central Level Public Interest Program for Scientific Research Institute (2016ZX310177 to N.W.), PUMC Youth Fund & the Fundamental Research Funds for the Central Universities (3332016006 to N.W.), Chinese Academy of Medical Sciences (CAMS) Initiative Fund for Medical Sciences (2016-I2M-3-003 to G.Q. and N.W., 2016-I2M-2-006 and 2017-I2M-2-001 to Z.W.), the Distinguished Youth Foundation of Peking Union Medical College Hospital (JQ201506 to N.W.), the 2016 PUMCH Science Fund for Junior Faculty (PUMCH-2016-1.1 to N.W.), and the National Key Research and Development Program of China (no. 2016YFC0901501 to S.Z.). This work was also supported by the Japan Agency for Medical Research and Development (AMED, number 17ek0109280h0001 and 17824969 to S.I.) and Japan Orthopedics and Traumatology Research Foundation (number 358 to K.T.), and the US National Institutes of Health, National Institute of Neurological Disorders and Stroke (NINDS R01 NS058529 and R35 NS105078 to J.R.L.), National Human Genome Research Institute/National Heart, Lung, and Blood Institute (NHGRI/NHLBI UM1 HG006542 to D.V. and J.R.L.), and the National Human Genome Research Institute (NHGRI K08 HG008986 to J.E.P.).
Author information
Authors and Affiliations
Consortia
Corresponding authors
Ethics declarations
Disclosure
J.R.L. has stock ownership in 23andMe, is a paid consultant for Regeneron Pharmaceuticals, and is a coinventor on multiple US and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, and bacterial genomic fingerprinting. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from the chromosomal microarray analysis and clinical exome sequencing offered in the Baylor Genetics Laboratory (http://bmgl.com). The other authors declare no conflicts of interest.
Additional information
These authors share senior authorship: Zhihong Wu, Shiro Ikegawa, James R. Lupski, Feng Zhang, Guixing Qiu.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Liu, J., Wu, N., Deciphering Disorders Involving Scoliosis and COmorbidities (DISCO) study. et al. TBX6-associated congenital scoliosis (TACS) as a clinically distinguishable subtype of congenital scoliosis: further evidence supporting the compound inheritance and TBX6 gene dosage model. Genet Med 21, 1548–1558 (2019). https://doi.org/10.1038/s41436-018-0377-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-018-0377-x
Keywords
This article is cited by
-
Molecular landscape of congenital vertebral malformations: recent discoveries and future directions
Orphanet Journal of Rare Diseases (2024)
-
Unraveling the genetic architecture of congenital vertebral malformation with reference to the developing spine
Nature Communications (2024)
-
Sequencing individual genomes with recurrent genomic disorder deletions: an approach to characterize genes for autosomal recessive rare disease traits
Genome Medicine (2022)
-
Delineation of dual molecular diagnosis in patients with skeletal deformity
Orphanet Journal of Rare Diseases (2022)
-
Expanding the mutation and phenotype spectrum of MYH3-associated skeletal disorders
npj Genomic Medicine (2022)