Abstract
Purpose
Pathogenic variants in KAT6A have recently been identified as a cause of syndromic developmental delay. Within 2 years, the number of patients identified with pathogenic KAT6A variants has rapidly expanded and the full extent and variability of the clinical phenotype has not been reported.
Methods
We obtained data for patients with KAT6A pathogenic variants through three sources: treating clinicians, an online family survey distributed through social media, and a literature review.
Results
We identified 52 unreported cases, bringing the total number of published cases to 76. Our results expand the genotypic spectrum of pathogenic variants to include missense and splicing mutations. We functionally validated a pathogenic splice-site variant and identified a likely hotspot location forde novo missense variants. The majority of clinical features in KAT6A syndrome have highly variable penetrance. For core features such as intellectual disability, speech delay, microcephaly, cardiac anomalies, and gastrointestinal complications, genotype– phenotype correlations show that late-truncating pathogenic variants (exons 16–17) are significantly more prevalent. We highlight novel associations, including an increased risk of gastrointestinal obstruction.
Conclusion
Our data expand the genotypic and phenotypic spectrum for individuals with genetic pathogenic variants in KAT6A and we outline appropriate clinical management.
Similar content being viewed by others
Introduction
Lysine (K) acetyltransferase 6 A (KAT6A, a.k.a. MOZ, MYST3) belongs to the MYST family of histone acetyltransferases that are defined by the presence of a highly conserved MYST domain consisting of acetyl-CoA binding motif and a zinc finger.1 The MYST family of proteins (KAT6A, KAT6B, KAT5, and KAT7) take part in a wide range of core cellular functions, such as chromatin remodeling, gene regulation, protein translation, metabolism, and cellular replication.2 Use of exome sequencing in patients with syndromic intellectual disability has revealed causative pathogenic variants in several genes that function as parts of chromatin remodeling complexes.3,4,5,6
De novo, rare, and protein-truncating genomic variants in a number of genes have been associated with cases of intellectual disability with speech delay.7,8 Genes involved in highly penetrant and syndromic developmental delay have high probability of loss-of-function intolerance (pLI) scores,9 a metric indicating that these same genes are not found to have predicted protein-truncating variants in a control population. Protein-truncating variants in two of the four MYST family genes, paralogs KAT6A (OMIM 6162680) andKAT6B (OMIM 606170 and 603736) have been associated with syndromic developmental delay. The phenotypic heterogeneity of pathogenic variants in both genes is striking. De novo truncating variants in KAT6B cause a spectrum of disorders, including genitopatellar syndrome (MIM 606170), Ohdo syndrome (Say–Barber–Biesecker–Young–Simpson SBBYS variant MIM 603736) and a Noonan syndrome–like disorder.10,11,12 Reported cases of KAT6A syndrome have been identified primarily through clinical or research exome sequencing in a gene-centric approach. A few cases have been identified through consortia exploring broad clinical phenotypes such as neutropenia.13
KAT6A and KAT6B each function in a multisubunit complex with three other proteins: BRPF1/2/3, ING5, and hEAF614. These proteins form a complex to acetylate lysine residues on histone H3 tails, thereby promoting a wide range of developmental programs. The importance of the KAT6A/B complex has been further highlighted by recently identified pathogenic variants in the binding partner, BRPF115,16 resulting in clinical features that overlap with syndromes caused by pathogenic variants in KAT6A and KAT6B.
The use of model organisms to investigate KAT6A function has provided insight into the role of KAT6A in vivo. Complete knockout mouse models result in embryonic lethality, due to a failure of hematopoiesis. A knock-in pathogenic variant that eliminates the KAT6A’s acetyltransferase function results in decreased life span, decreased body weight, and proliferation defects.17 Tissue- or cell-specific knockout has shown that KAT6A regulates transcriptional programs important for skeletogenesis, hematopoiesis, and splenic and thymic function.17,18,19 Further studies demonstrated that KAT6A-mediated acetylation promotes memory B-cell formation and the CD8 T-cell response to viral infection.20,21 Transcriptomic profiles of human fibroblast cell lines derived from patients harboring heterozygous KAT6A truncating pathogenic variants demonstrated altered expression of p53-associated genes.4
To date, eight papers4,5,6,13,22,23,24,25 describe 24 patients with pathogenic variants in KAT6A. In this paper, we add 52 novel and comprehensively phenotyped cases and review all previously published cases.
Materials and methods
Research cohort
Phenotypic information from patients with likely causativeKAT6A pathogenic variants was obtained by three methods: through clinical geneticists, through an online survey to families, and through literature review. The cohorts were independently identified and therefore some individuals were identified through two methods. In these cases data were combined. All patients/families included in the study provided consent through the treating clinician or through the institutional review board–approved patient/family survey. Written consent for publication of patient photos was also obtained.
In the first cohort (N = 33), information was obtained from the primary clinical geneticist using a targeted phenotypic questionnaire designed to identify a spectrum of clinical phenotypes in patients with convincing de novo pathogenic variants in KAT6A. In this cohort some patients were identified through the Deciphering Developmental Disorders (DDD) Study, and the DDD Complementary Analysis Project allowed us to access initial phenotypic data.7 Additional patients in Australia, Holland, Japan, Finland, Norway, and the United States were identified through communication of the lead authors with treating physicians and KAT6A variants were identified through clinical or research exome sequencing.
A second cohort (N = 43) was recruited through social media and patient advocacy groups. This was performed independently. We collected information allowing for us to match cases. A parent or family member was asked to complete an online survey spanning birth history, developmental milestones, current treatments, and associated medical conditions. Detailed information about the clinically identified genetic change in KAT6A gene was also collected.
A third cohort (N = 24) was identified from the published medical literature. Approximately 40% of families where the proband case was previously published provided updated information via online survey.
Splice-site variant analysis
RNA was extracted from peripheral blood samples and underwent reverse transcription using a High Capacity cDNA RT kit (Thermo Fisher Scientific). Polymerase chain reaction (PCR) amplification was performed using custom designed primers (see Supplemental information for additional details) and Sanger sequencing was performed using ABI3730 DNA sequencer (Applied Biosystems).
Statistical analysis
To assess significant differences between our early- and late-truncating variant cohorts, we performed a two-tailed Fisher's exact test26 to determine if the differences within groups were significant.
Results
Our study comprised a total of 75 patients with pathogenic or likely pathogenic variants in the gene KAT6A and 1 patient with a variant of unknown significance (VUS). Of these, 70% (52/76) are novel cases that have not been previously reported in the literature. Their ages range from 1 to 32 years of age and the cohort is 49% female and 51% male.
Genotype analysis
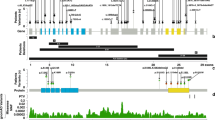
Within the 52 novel cases, we identified 44 novel genetic variants, of which 88% (39/44) are predicted to result in a truncating frameshift or nonsense variant. Some locations have recurrent truncating changes, primarily located in the acidic domain, which is rich in arginine residues (amino acid positions 1019, 1024, and 1129).
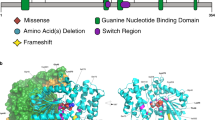
Five novel pathogenic variants are missense changes at highly conserved residues (Supplemental Fig. 2). Four of the five missense changes have been classified as likely pathogenic because they are de novo, rare,9 match the clinical phenotype, and are predicted to be deleterious based on in silico algorithms.27,28,29 The remaining missense change, p.S371Y, is classified as a VUS as the patient fulfilled the criteria above except the variant was ultimately found to be maternally inherited from an unaffected parent. Although the missense Z-score9 for KAT6A is nonsignificant at Z = 2.14, this score represents a depletion across the entire gene and does not account for regional variation. In our missense cases, three of the individuals have de novo missense changes within a highly conserved region and are known to bind to RUNX1/2, a gene important in transcriptional transactivation (Fig. 1c).
KAT6A domains and the distribution of pathogenic genetic variants.a We observed a total of 76 patients with 61 unique genetic variants across the 2004–amino acid protein. The pathogenic variants in new patients are shown above the gene; previously reported variants are displayed below the gene. Missense pathogenic variants are denoted in red while protein-truncating pathogenic variants are denoted in black font. Protein ID for KAT6A is NP_006757.2 and various protein domains are: NEMM domain (AA 1–206), PHD domains (AA 207–313), HAT domain (AA 314–787), acidic domain (788–1414), and Ser/Met domain (1414–2004). b Splice changes are denoted in blue demonstrating their approximate intronic location in the gene model. c Missense changes are located near important functional domains of KAT6A described in UniProt.34d The functional effect of the identified splice variant in patient 17 was validated from sequencing of the complementary DNA (cDNA) product of blood RNA. The splicing effect resulted in a deletion of eight base pairs and a frameshift change. WT wild type
We have identified 4 individuals with predicted splice-site changes due to substitutions in canonical splice site of KAT6A exons. For patient 17 with pathogenic variant at c.1364-2 A>T, sequencing of KAT6A cDNA from a patient blood sample demonstrated that the splice variant resulted in an 8-bp deletion in exon 8, which is predicted to cause protein truncation (Fig. 1d).
Clinical features of newly reported patients
Developmental delay and intellectual disability
Intellectual disability and developmental delay are universal. Intellectual disability (ID) varies from mild to severe. One young adult has a driver's license. Hypotonia is common and contributes to motor delay. Often, truncal hypotonia was associated with limb hypertonia; this was more notable in the neonatal period. Specific speech and language delay combined with motor delay may lead to an overestimate of ID in the early years. To date no individuals have been reported without ID though this may represent a selection bias because most patients have been identified by sequencing patients with ID.
A correlation is observed between the site of the pathogenic variant and the level of ID. In cases in which the level of ID was reported on, 95% of late-truncating cases (exon 16 and 17) were rated as moderate or severe, while 60% of early-truncating cases (exons 1–15) were rated as mild (Supplementary Fig. 3). This pattern was also observed when we considered the age of achieving developmental milestones for affected patients. A previously reported patient with a full-gene deletion had a mild ID (20).
Oromotor dyspraxia
Marked expressive speech delay is universal. Many struggle with articulation. Speech delay is often described as a form of verbal dyspraxia. Receptive language is consistently more developed than expressive language. One patient with mild ID achieved a score typical for receptive language for his age. Use of sign language and communication aids are helpful. Communication difficulties are a source of frustration. Despite significant delays many children do make progress. For example, a patient who spoke four words at the age of 4 now speaks fluently as an adult. However, there are individuals with KAT6A syndrome who remain nonverbal into adulthood.
Feeding difficulties are commonly associated with verbal dyspraxia due to oromotor dysfunction. Seventy-eight percent of patients experienced feeding difficulties. Many patients had difficulty establishing feeding at birth and nasogastric feeding was often required. In addition, several patients have dysphagia and a few have had recurrent aspirations.
Other gastrointestinal problems
The high prevalence of reflux and constipation is suggestive of dysfunctional intestinal motility. Reflux is a significant issue, and while some outgrow this a few report persistent vomiting and retching well beyond infancy. Many patients have had a gastrostomy tube for feedings and small number underwent a fundoplication. Constipation is a significant issue for over half of our patients and many are on long-term laxatives. In addition, four patients in this study (patients 6, 22, 42, and 46) had bowel obstruction. One required surgery for a duodenal web and malrotation, the second also required surgery for a malrotation, the third had a small bowel obstruction leading to the resection of part of the ileum, and the fourth required three laparotomies for a bowel obstruction, and subsequently experienced gastrointestinal failure and was transferred to hospice care. This patient also had anal stenosis and gastroesophageal nerve impairment. Interestingly malrotation and duodenal rupture has been reported inKAT6B related disease30 suggesting shared underlying mechanisms.
Cardiac malformations
Cardiac malformations are present in half (51%) of our cohort. Most frequent are septal defects including atrial septal defects (34%), ventricular septal defects in (8%), and persistence of the fetal anatomy (19%) (patent foreman ovale, and persistent ductus arteriosus). At least 45% of patients with cardiac malformations required intervention (open heart surgery or via cardiac catheterization). A minority of our patients are awaiting cardiology review and have not yet had an echocardiogram. The prevalence of cardiac lesions highlights the need for early cardiology assessment.
Hematological and immunological associations
While the majority of patients did not report hematological or immunological deficiencies, three individuals report isolated moderate to severe neutropenia.5,13 Many patients' parents reported frequent infections, and some were reported to take longer to recover from infections compared with peers. The majority, however, did not report that they experienced frequent infections and those who did tended to report common childhood illnesses including otitis media and upper and lower respiratory tract infections. Many children experience recurrent infections of these types, so this is not necessarily indicative of immunodysfunction.
Some unusual infections have been noted. Patient 7 has recurrent and extensive herpes simplex infections of the face and eyes and patient 13 has had impetigo and a separate staph infection. Patient 2 also has a B-cell and T-cell immunodeficiency, hypogammaglobulinemia for which she receives regular intravenous immunoglobulins, in addition to episodes of suspected perianal streptococcal dermatitis. Patient 20 has hypogammaglobulinemia. Patient 11 mounted a low immune response to HibB and pneumococcal vaccine requiring booster vaccines. Patient 1 has unexplained persistent thrombocytopenia. One hypothesis is that KAT6A syndrome may result in a variety of abnormalities of blood cell lines. Alternately, there may be other genetic or environmental factors in each individual.31 Additional research is required to define this further. Structural abnormalities can also predispose to infection. Patients 3 and 10 have renal abnormalities and recurrent urinary tract infections.
Facial features
A broad nasal tip, which may become more obvious with age, and a thin, tented upper lip, are the most consistent facial features in patients with KAT6A syndrome. Other common facial features include bitemporal narrowing, prominence of the nasal bridge, and a short and flat philtrum. Notable features present in a significant minority are epicanthic folds and low-set and posteriorly rotated ears, which are occasionally folded (Fig. 2).
Within the mouth a high arched narrow palate was noted in a few patients and teeth abnormalities were common. Abnormal peg-shaped teeth have been reported previously4,5 and are also seen in a number of our newly reported patients. Other dental abnormalities reported are small tooth size, supernumary teeth, and dental crowding. Cleft palates are not frequently seen (reported in two patients).
Skull and brain abnormalities
Many patients with KAT6A syndrome have had magnetic resonance image (MRI) scans of the brain and major structural anomalies are rare. One patient has been reported with a pituitary malformation and related hormone deficiencies.25 No other pituitary stalk abnormalities have been reported. Some mild abnormalities have been reported including a thin corpus callosum or delayed myelination that resolves over time. Other structural anomalies observed in our cohort include a large cisterna magna in patient 1, a variant venous anatomy including anomalous venous sinus that traverses the falx cerebri in patient 26, and hydrocephalus and a Chiari malformation in patient 33.
Craniosynostosis is reported in a total of six patients.5 Initially, microcephaly was reported in 33% of previously published patients4,5,6 and in our larger cohort, only 25% individuals had microcephaly, which was not always present at birth. Seizure activity has only been reported in seven patients and there is no consistency in seizure type (see Supplementary Table 2).
Eye features
Strabismus is reported in 54% of patients. This can be intermittent and is often worse when fatigued. Some patients have been treated with patching or surgery. Strabismus can result in amblyopia and this is more likely to occur if it is unrecognized and untreated. Amblyopia is reported in several patients.
Refractory errors are reported in a minority of patients. Myopia is more commonly reported than hypermetropia. Delayed visual maturation, cortical visual impairment, and astigmatism are also occasionally reported as well as isolated cases of photophobia (patient 9), latent horizontal nystagmus (patient 11), and Jeavons epilepsy (patient 47).
Behavioral issues
An increased frequency of behavioral difficulties is noted in our cohort. Autism and autistic features have been reported in approximately 25% of newly reported cases. Temper tantrums, inappropriate laughing, hand flapping, and increased anxiety are also mentioned in multiple patients. The majority of patients in our cohort do not report behavioral difficulties and many are described as being good natured, happy, and sociable.
Sleep
Over 30% of patients in this cohort reported sleep disturbance. This included difficulty initiating and maintaining sleep, and five patients reported central obstructive sleep apnea.
Other clinical features and clinical guidelines
A summary of all clinical features described is located in Table 1. Supplementary Tables 2 and 3 contain further details on unique features seen in a few patients. For example, we found a small number of patients had undescended testes (boys), clinodactyly, and/or brachydactyly.
Based on the features described above, we have put together a set of general guidelines for clinicians to help guide the clinical workup for patients with a pathogenic genetic variant in KAT6A (Table 2).
Phenotypic differences between early and late-onset truncating pathogenic variants
We further subdivided the cases between early-truncating pathogenic variants (exons 1–15) and late-truncating pathogenic variants (exons 16–17) to determine if there was a difference in severity of phenotype or prevalence of specific syndromic features. Across 19 features (Table 1) we performed Fisher's exact test and identified 8 subphenotypes that were significantly more common in patients with late-truncating pathogenic variant (Fig. 3c). These included microcephaly, neonatal hypotonia, feeding difficulties, reflux, constipation, congenital hearing defects, and frequent infections. Our data on the subphenotypes fit with the trend we observed, in which there is more severe global developmental delay and intellectual disability in patients with late-truncating pathogenic variants.
Developmental delay, intellectual disability, and early and late-truncating pathogenic variant.a Developmental milestones in patients with pathogenic variants in KAT6A. Delay in childhood milestones are commonly seen, with milder delays in social and motor milestones and more severe delay noted in acquisition of verbal language.b Intellectual disability and developmental delay are more severe for truncating pathogenic variant in last two exons. A. More severe intellectual disability is more commonly seen in patients with truncating pathogenic variants in the last two exons of KAT6A as compared with early-truncating pathogenic variants (in exons 1–15). Contributing clinicians were asked to rate the level of intellectual disability as mild, moderate, or severe on a scale from 1 to 3 (1 = mild to 3 = severe). Patients collected through the survey or through the Deciphering Developmental Disorders (DDD) Study were asked to report the age when key developmental milestones were reached. We rated the severity of delay for four milestones as mild, moderate, or severe: first smile (mild = 2–4 months, mod = 4–6 months, severe = 6 months+); sitting (mild = 6–12 months, mod = 12–18 months, severe = 18 months+); walking (mild 12–24 months, mod = 24–30 months, severe = 30 months+); first word (mild = 1–3 years, mod = 3–5 years, severe = 5 years+). The scores for each individual were then averaged to give a score of 1 to 3. c Several syndromic features were seen less commonly in patients with early-truncating pathogenic variants . This includes microcephaly, neonatal hypotonia, gastrointestinal complications, and congenital heart defects.
Discussion
The mutational spectrum and the wide age range of patients allowed us to perform a comprehensive phenotypic assessment to elucidate the clinical phenotypes in childhood, adolescence and adulthood.
The wide phenotypic spectrum in individuals with KAT6A variants highlights the continued importance of exome sequencing to identify the genetic etiology for patients with syndromic ID.3,7 Patients with KAT6A pathogenic variant s were not phenotypically grouped prior to the advent of clinical exome sequencing as there are no uniqueand unifying features that allowed for easy recognition by physicians. While patients with KAT6A syndrome do share many phenotypic features, many of these are common to a wide range of developmental syndromes. Accurate and detailed reporting of the phenotype is critical for affected families. KAT6A is one of the more common causes of undiagnosed syndromic intellectual disability7 with some reports suggesting a rate as high as 1% of undiagnosed syndromic developmental delay.4
Prior to this report, the majority of pathogenic variants were found to be de novo, truncating, and located in the ultimate and penultimate exons of the gene, which make up over half of the protein. Our analysis identifies hotspot nonsense pathogenic variants within the penultimate exons at amino acid positions 1019, 1024, and 1129 that account for 19.1% (13/68) of pathogenic variants in unrelated individuals. Protein-truncating variants have been identified throughout the length of the gene, and the number of cases we present in this article allows us to consider the genotype–phenotype correlations. We observe a bias of increased severity of developmental delay and an increased frequency of microcephaly, hypotonia, cardiac anomalies, and gastrointestinal complications associated with truncating pathogenic variants in the last two exons. This suggests a potential role for nonsense mediated decay (NMD), where truncating pathogenic variants in the first 15 exons trigger NMD mechanisms and result in haploinsufficiency while pathogenic variants s in exons 16 and 17 would not result in NMD, therefore the messenger RNA (mRNA) would result in a translated but dysfunctional protein that may have gain-of-function or dominant negative effects.
It should be noted that assessment of intellectual disability and developmental delay was based on clinicians rating the level of the patient’s intellectual disability as mild, moderate, or severe rather than from formal IQ testing. However, it is unlikely that this resulted in significant bias because this effect was not predicted before collecting the patient data. A large fraction of patients with KAT6A syndrome have had some form of biochemical and metabolic testing as a part of the clinical genetic workup. None of these cases demonstrated a clear and consistent metabolic dysfunction, by standard clinical biochemical genetic testing.
The role of nonsense mediated decay allowing for differing mechanisms of pathogenic variants within the same gene has been observed in pathogenic variants of related gene KAT6B, which results in two distinct syndromes, SBBYS and genitopatellar syndrome.10,11 It has been postulated that differential truncating pathogenic variants in exon 18 may have a gain-of-function or even dominant negative effects.10 The molecular effects of different pathogenic variant localization remain to be validated in careful functional studies.
The lack of distinctive clinical features makes the attribution of KAT6A syndrome to missense variants in KAT6A particularly challenging. In our study, we have six patients with missense variants (including one previously reported case), five of which are de novo and one maternally inherited. While all of these variants fall into highly conserved regions of the protein (Supplemental Fig. 2) further functional studies are needed to confirm pathogenicity. Although our sample size is small, patients with missense variants have not, to date, shown a cardiac phenotype. We are aware that there are many more individuals with de novo missense reported through the DDD study, for which the pathogenicity is unclear. Some of these individuals have multiple potentially pathogenic variants further increasing the difficulty of assigning pathogenicity. Additionally, it is becoming increasingly evident that multiple variants may contribute to the phenotype; however it remains difficult to assess the relative contribution and interaction of multiple variants within an individual.31,32
We note significant clinical variability in our cohort. This is not surprising as KAT6A functions as an epigenetic modifier, so its molecular effects are more nuanced and influenced by both background genetic variation and the environment. While we have a fairly large cohort, it is not large enough to confidently identify rare associations. For instance, we cannot yet be certain about the link with KAT6A syndrome and immunodeficiency or pituitary anomalies. When individuals with pathogenic KAT6A variants are found to have unusual features they should be further assessed for either a second pathogenic variant,31 or nongenetic cause, in addition to considering the possibility that the KAT6A pathogenic variant is responsible.
An interesting aspect of our study is that we collected data for many of our patients through an online family survey. The family questionnaire was received warmly and many families responded in a short timeframe. It proved to be a useful, relatively low resource, and efficient way to collect data. It allowed us to collect updated information for patients who had been previously reported in the literature. It should be noted however that although many individuals started the survey, only 60% completed the survey and were included in our study. It is not surprising that the patient families are heavily invested in their care. Many maintained a deep working knowledge of their child’s medical condition. Collecting information from the family relies on the engagement of the families and a basic understanding of medical terminology, therefore consulting with family members to ensure accessibility of survey questions is critical to designing a successful survey. The family survey and the clinician questionnaire had to be designed with different wording to take into account the different baseline level of medical knowledge of the two groups. In the family survey, parents were asked to select keywords that the child was noted to have by a physician (e.g., wide-spaced nipples, small jaw, small head, abnormal teeth, etc.). Collecting information from family members about dysmorphic facial features can be offensive and family members are not trained to assess this. We therefore also requested families to share photos so that this could be assessed by trained dysmorphologists. We were able to obtain a photo from 9 of the 15 survey-only individuals. Other studies have found family questionnaires an effective way of investigating disease phenotypes.33 As the number of genetic syndromes increases, utilization of well-designed clinical surveys can provide invaluable data for clinical definition and for identifying a baseline by which future therapies can be measured.
Our report is the most comprehensive phenotypic evaluation of patients with pathogenic variants in KAT6A. We have provided significant insight into the range of phenotypic expressivity observed in patients with KAT6A syndrome. Further functional studies assessing the role of genetic pathogenic variantsare required to understand the effect of various missense pathogenic variants and how early and late-truncating pathogenic variants s ultimately affect downstream molecular processes.
Change history
19 August 2020
The original version of this Article did not include Tatjana Bierhals, Maja Hempel, Jessika Johannsen & Davor Lessel in the list of authors for their contribution to data collection and analysis. This has now been corrected in both the PDF and HTML versions of the Article.
19 August 2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
Avvakumov N, Cote J. The MYST family of histone acetyltransferases and their intimate links to cancer. Oncogene. 2007;26:5395–5407.
Voss AK, Collin C, Dixon MP, Thomas T. Moz and retinoic acid coordinately regulate H3K9 acetylation, Hox gene expression, and segment identity. Dev Cell. 2009;17:674–686.
Lee H, Deignan, JL, Dorrani, N, Strom, SP, Kantarci, S, Quintero-Rivera, F, et al. Clinical Exome Sequencing for Genetic Identification of Rare Mendelian Disorders. JAMA. 2014;312:1880–1887.
Arboleda VA, Lee H, Dorrani N, et al. De novo nonsense mutations in KAT6A, a lysine acetyl-transferase gene, cause a syndrome including microcephaly and global developmental delay. Am J Hum Genet. 2015;96:498–506.
Tham E, Lindstrand A, Santani A, et al. Dominant mutations in KAT6A cause intellectual disability with recognizable syndromic features. Am J Hum Genet. 2015;96:507–513.
Millan F, Cho MT, Retterer K, et al. Whole exome sequencing reveals de novo pathogenic variants in KAT6A as a cause of a neurodevelopmental disorder. Am J Med Genet A. 2016;170:1791–1798.
Wright CF, Fitzgerald TW, Jones WD, et al. Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet. 2015;385:1305–1314.
Deciphering Developmental Disorders Study. Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438.
Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291.
Campeau PM, Lu JT, Dawson BC, et al. The KAT6B-related disorders genitopatellar syndrome and Ohdo/SBBYS syndrome have distinct clinical features reflecting distinct molecular mechanisms. Hum Mutat. 2012;33:1520–1525.
Clayton-Smith J, O’Sullivan J, Daly S, et al. Whole-exome-sequencing identifies mutations in histone acetyltransferase gene KAT6B in individuals with the Say-Barber-Biesecker variant of Ohdo syndrome. Am J Hum Genet. 2011;89:675–681.
Szakszon K, Salpietro C, Kakar N, et al. De novo mutations of the gene encoding the histone acetyltransferase KAT6B in two patients with Say-Barber/Biesecker/Young-Simpson syndrome. Am J Med Genet A. 2013;161A:884–888.
Gauthier-Vasserot A, Thauvin-Robinet C, Bruel AL, et al. Application of whole-exome sequencing to unravel the molecular basis of undiagnosed syndromic congenital neutropenia with intellectual disability. Am J Med Genet A. 2017;173:62–71.
Yang XJ. MOZ and MORF acetyltransferases: molecular interaction, animal development and human disease. Biochim Biophys Acta. 2015;1853:1818–1826.
Mattioli F, Schaefer E, Magee A, et al. Mutations in histone acetylase modifier BRPF1 cause an autosomal-dominant form of intellectual disability with associated ptosis. Am J Hum Genet. 2017;100:105–116.
Yan K, Rousseau J, Littlejohn RO, et al. Mutations in the chromatin regulator gene BRPF1 cause syndromic intellectual disability and deficient histone acetylation. Am J Hum Genet. 2017;100:91–104.
Perez-Campo FM, Borrow J, Kouskoff V, Lacaud G. The histone acetyl transferase activity of monocytic leukemia zinc finger is critical for the proliferation of hematopoietic precursors. Blood. 2009;113:4866–4874.
Sapountzi V, Cote J. MYST-family histone acetyltransferases: beyond chromatin. Cell Mol Life Sci. 2011;68:1147–1156.
Thomas T, Corcoran LM, Gugasyan R, et al. Monocytic leukemia zinc finger protein is essential for the development of long-term reconstituting hematopoietic stem cells. Genes Dev. 2006;20:1175–1186.
Good-Jacobson KL, Chen Y, Voss AK, Smyth GK, Thomas T, Tarlinton D. Regulation of germinal center responses and B-cell memory by the chromatin modifier MOZ. Proc Natl Acad Sci USA. 2014;111:9585–9590.
Newman DM, Sakaguchi S, Lun A, et al. Acetylation of the Cd8 locus by KAT6A determines memory T cell diversity. Cell Rep. 2016;16:3311–3321.
Chisei Satoh RM, Akira Kinoshita, Hiroyuki Mishima, Michiko Doi, Mutsuko Miyazaki, Masafumi Fukuda, et al. Three brothers with a nonsense mutation in KAT6A caused by parental germline mosaicism. Human Genome Variation. 2017;4:17045.
Elenius V, Lahdesmaki T, Hietala M, Jartti T. Food allergy in a child with de novo KAT6A mutation. Clin Transl Allergy. 2017;7:19.
Murray CR, Abel SN, McClure MB, et al. Novel causative variants in DYRK1A, KARS, and KAT6A associated with intellectual disability and additional phenotypic features. J Pediatr Genet. 2017;6:77–83.
Zwaveling-Soonawala N, Maas SM, Alders M, et al. Variants in KAT6A and pituitary anomalies. Am J Med Genet A. 2017;173:2562–2565.
Fisher RA On the interpretation of χ2 from contingency tables, and the calculation of P. J Royal Stat Soc. 1922;87–94.
Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315.
Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081.
Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249.
Lonardo F, Lonardo MS, Acquaviva F, Della Monica M, Scarano F, Scarano G Say-Barber-Biesecker-Young-Simpson syndrome and Genitopatellar syndrome: Lumping or splitting? Clin Genet. 2017;1–9.
Posey JE, Harel T, Liu P, et al. Resolution of disease phenotypes resulting from multilocus genomic variation. N Engl J Med. 2017;376:21–31.
Katsanis N. The continuum of causality in human genetic disorders. Genome Biol. 2016;17:233.
Wang RT, Silverstein Fadlon CA, Ulm JW, et al. Online self-report data for Duchenne muscular dystrophy confirms natural history and can be used to assess for therapeutic benefits. PLoS Curr. 2014;6. https://doi.org/10.1371/currents.md.e1e8f2be7c949f9ffe81ec6fca1cce6a.
UniProt Consortium T. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45(D1):D158-D169.
Acknowledgements
The authors would like to acknowledge the KAT6A Foundation (http://www.kat6a.org/) and all the patients and their families who completed the survey and consented to this study. We would also like to acknowledge the DDD study. The DDD study presents independent research commissioned by the Health Innovation Challenge Fund (grant number HICF-1009-003), a parallel funding partnership between the Wellcome Trust and the Department of Health, and the Wellcome Trust Sanger Institute (grant number WT098051). The views expressed in this publication are those of the author(s) and not necessarily those of the Wellcome Trust or the Department of Health. The study has UK Research Ethics Committee (REC) approval (10/H0305/83, granted by the Cambridge South REC, and GEN/284/12 granted by the Republic of Ireland REC). The research team acknowledges the support of the National Institute for Health Research (NIHR), through the Comprehensive Clinical Research Network. This study makes use of DECIPHER (http://decipher.sanger.ac.uk), which is funded by the Wellcome Trust. J.K. held an NIHR Academic Clinical Fellowship post during this course of this study. This work was also supported by an NIH Early Independence Award to V.A.A. (DP5OD024579), and the KAT6A Foundation.
Disclosure
The authors declare no conflicts of interest.
Author information
Authors and Affiliations
Consortia
Corresponding authors
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Kennedy, J., Goudie, D., Blair, E. et al. KAT6A Syndrome: genotype–phenotype correlation in 76 patients with pathogenic KAT6A variants. Genet Med 21, 850–860 (2019). https://doi.org/10.1038/s41436-018-0259-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-018-0259-2
Keywords
This article is cited by
-
The omics era: a nexus of untapped potential for Mendelian chromatinopathies
Human Genetics (2024)
-
Epigenetics of cognition and behavior: insights from Mendelian disorders of epigenetic machinery
Journal of Neurodevelopmental Disorders (2023)
-
Luteolin promotes KAT6A gene expression
Journal of Rare Diseases (2023)
-
KAT6A mutations in Arboleda-Tham syndrome drive epigenetic regulation of posterior HOXC cluster
Human Genetics (2023)
-
Chromatin regulators in the TBX1 network confer risk for conotruncal heart defects in 22q11.2DS
npj Genomic Medicine (2023)