Abstract
Purpose
To systematically investigate the longer-term clinical and health economic impacts of genomic sequencing for rare-disease diagnoses.
Methods
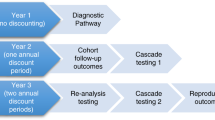
We collected information on continuing diagnostic investigation, changes in management, cascade testing, and parental reproductive outcomes in 80 infants who underwent singleton whole-exome sequencing (WES).
Results
The median duration of follow-up following result disclosure was 473 days. Changes in clinical management due to diagnostic WES results led to a cost saving of AU$1,578 per quality-adjusted life year gained, without increased hospital service use. Uninformative WES results contributed to the diagnosis of non-Mendelian conditions in seven infants. Further usual diagnostic investigations in those with ongoing suspicion of a genetic condition yielded no new diagnoses, while WES data reanalysis yielded four. Reanalysis at 18 months was more cost-effective than every 6 months. The parents of diagnosed children had eight more ongoing pregnancies than those without a diagnosis. Taking the costs and benefits of cascade testing and reproductive service use into account, there was an additional cost of AU$8,118 per quality-adjusted life year gained due to genomic sequencing.
Conclusion
These data strengthen the case for the early use of genomic testing in the diagnostic trajectory, and can guide laboratory policy on periodic WES data reanalysis.
Similar content being viewed by others
Introduction
A growing body of evidence is providing support for the use of clinical genomic sequencing early in the diagnostic trajectory of patients with rare genetic diseases. Early genomic testing of children has high diagnostic and clinical utility1,2 and is cost-effective compared with the cost of the current standard diagnostic pathway.3,4,5 However, a number of uncertainties remain around the use of genomic sequencing in clinical care.
First, the longer-term impact of diagnostic genomic results on patient management, health-care service provision and family decision-making is not known. Rare diseases are commonly perceived either as “untreatable” or requiring expensive therapies that may impact on constrained health-care systems. Anecdotal evidence from families highlights the benefits of putting an end to the “diagnostic odyssey,” targeting treatment and enabling reproductive planning,6 but to date studies have predominantly looked at the immediate impact on care,1,2,4,7,8,9,10 and only one study has examined downstream patient care and cost consequences over a period of a year.11
Second, the impact of “uninformative” results on further investigation and health-care activity is poorly understood. An uninformative result could either lead to further investigation and health-care activity focused on achieving a diagnosis or, alternatively, diminished diagnostic efforts. Stored genomic data provide unique opportunities for reanalysis in the light of new gene discoveries, improved bioinformatics techniques, and changes in clinical circumstances. Guidelines propose that storage and reanalysis is good practice,12 and data from several studies indicate diagnostic effectiveness.13,14,15 However, laboratory practices vary from not offering reanalysis to automated reanalysis at no additional charge to reanalysis on clinician request at defined time intervals and at a cost.16 The cost-effectiveness and clinical impact of reanalysis compared with ongoing standard investigation (e.g., candidate gene testing) have not been determined, but are necessary to inform clinical and laboratory policies.
Finally, cost-effectiveness studies of longer-term outcomes are lacking. We previously reported a cost-effectiveness analysis of singleton whole-exome sequencing (WES) compared with standard diagnostic care in a prospective clinical cohort of infants with features suggestive of monogenic disorders. We found that when WES is used as a first-tier sequencing test, there is a saving of AU$2182 (US$1702) per additional diagnosis compared with the standard diagnostic pathway.1,3 While there are a small number of studies on the cost-effectiveness of WES, all of which support the early use of genomic testing to improve cost-effectiveness,3,4,5,17,18,19,20 no studies have taken into account the health outcomes at follow-up, which are typically measured as additional quality-adjusted life years (QALYs). Cost per QALY is the preferred metric to inform resource allocation in health-care systems. As a guide, the cost per QALY threshold used by the UK’s National Institute for Health and Care Excellence, over which interventions are less likely to be recommended for funding, is £20,000–30,000 (AU$33,757–50,636).
Systematic investigation of the long-term clinical and cost impacts of genomic sequencing is needed to guide its implementation into health systems and services. Health system funders, in particular, need evidence for policy, resource allocation, and service planning. We addressed this gap by following up the cohort of infants with suspected monogenic disorders who underwent singleton WES early in the clinical trajectory from our earlier study.1,3 In the current study, we analyze the broader clinical impacts and cost-effectiveness of informative and uninformative results on continuing diagnostic investigation, changes in management, cascade testing in first-degree relatives, and parental reproductive planning and outcomes. Our results show the broader longer-term effectiveness and cost-effectiveness of genomic sequencing in clinical care.
Materials and methods
Study design
Infants with suspected monogenic disorders were recruited from a single tertiary pediatric center (Royal Children’s Hospital, Melbourne, Australia). Recruitment was prospective and is fully described elsewhere.1 Briefly, infants from newborn to two years of age were eligible if they presented with multiple congenital abnormalities and dysmorphic features, or other features strongly suggestive of monogenic disorders, e.g., neurometabolic conditions and skeletal dysplasias. Singleton WES was performed early in the diagnostic trajectory as a first-tier sequencing test. Analysis of the WES data was limited to genes known to cause monogenic disorders (the “Mendeliome”; https://www.vcgs.org.au/sites/default/files/downloads/Clinical%20Exomes%20Gene%20List_0.pdf), and only variants relevant to the participant’s phenotype were assessed with regards to pathogenicity. The gene content of the “Mendeliome” was updated every 6 months, with data from “unsolved” patients reanalyzed at the time of each update.
Cost-effectiveness analyses were performed at follow-up to examine the improvements in health-related QALYs gained for the children and their parents resulting from changes in patient management and reproductive service utilization. The cost-effectiveness analysis was undertaken from the payer's (funded hospital system) perspective.
The study was part of the Melbourne Genomics Health Alliance Demonstration Project17 and received Human Research Ethics Committee approval (13/MH/326). Informed written consent was obtained from the parents of the participants.
Data collection
Patient follow-up data were collected between February 2014 and October 2016, such that patients were followed up for a minimum of 12 months after WES result disclosure. A clinical geneticist (Z.S.) extracted information on patient outcomes, including survival, changes in management, diagnostic investigations, tertiary pediatric hospital use, cascade testing in family members, and reproductive outcomes from the medical records held at the Royal Children’s Hospital, Melbourne and the Victorian Clinical Genetics Services, Melbourne, as well as from referring clinicians. Data on patient health states were collected via a parent survey, with child health states verified by a clinical geneticist (Z.S.). Specific health-related utility values for each child’s health state were assigned using utilities based on parent-reported preferences for health states in young children.21 Where possible, we used patient-reported survey data to assign a health state to a child, with clinical opinion being utilized when this was not available.
We obtained the costs of investigations, medications, and patient encounters from the hospital, state government, and testing laboratories. Costs in overseas currencies were converted into Australian dollars at the time when accounts were paid.
Statistical analysis
Descriptive statistics were used to describe the health-care utilization of tested infants; i.e., length of follow-up, number of hospital admissions, days in hospital and number of tertiary outpatient appointments (general pediatrics, pediatric subspecialist, and clinical genetics). Total, pre- and post-test hospital utilization was analyzed. Dot plots were used to visualize the distribution of variables to determine whether to use parametric or non-parametric measures. Means, standard deviation, CIs between means, and differences between means were determined for normally distributed variables. For non-normally distributed variables, medians and interquartile ranges were calculated, and CIs for the differences in medians were determined using quantile regression. Differences in hospital utilization were investigated using two-sided t-tests for normally distributed variables and the Wilcoxon rank-sum test for non-normally distributed variables. All analyses were performed using Stata 11.0 (Stata, College Station, TX, USA).
Health economic analysis
Cost-effectiveness of WES data reanalysis
We evaluated two service delivery models representing the re-examination of data from unsolved cases with ongoing suspicion of monogenic disorder at different time intervals—namely, once in 18 months (model 1) and once every 6 months for 18 months (model 2), and compared these with ongoing standard diagnostic care.
The cost of reanalysis was AU$350 per patient, with an additional cost of AU$300 if new diagnostic findings were reported. We calculated the cost per patient, cost per diagnosis, and incremental cost per additional diagnosis for all three diagnostic pathways.
Cost–utility analysis of the changes in patient management and reproductive service utilization
We conducted cost–utility analyses of genomic testing using WES and subsequent changes in patient management and reproductive service utilization at follow-up based on: (1) changes in costs and health outcomes (QALYs) due to changes in clinical management in children only; and (2) changes in costs and health outcomes (QALYs) due to changes in clinical management, cascade testing, and reproductive outcomes in children and their families (first-degree relatives). A QALY is a metric used to measure the utility of a health intervention by quantifying the relative value of an additional year of life in a particular state of health. Full health is valued at 1 and death is valued at 0. Results were presented as an incremental cost-effectiveness ratio (ICER), which is the difference in cost between the standard pathway and the intervention, divided by the difference in their effect in terms of QALYs gained.
To examine the uncertainties of the ICERs, we created 1000 replicated datasets using bootstrap simulation and estimated ICER for each replicated dataset generating a distribution of the ICER. We estimated 95% confidence intervals (CIs) for ICERs based on their distributions using the percentile method. Analyses were performed using SAS version 9.4 (https://www.sas.com/en_au/software/sas9.html).
Key underlying assumptions in modeling
It was assumed that attitudes toward family planning were the same across diagnosed and undiagnosed groups, with the additional reproductive confidence provided by molecular diagnosis being the factor that influenced additional births. For diagnosed patients, QALYs gained were based on clinicians’ prognosis of disease progress in the absence of a diagnosis, and thus no change in management. In the case of reproductive outcomes, child health utility was estimated based on health condition at the time of birth, with full utility for a healthy child born (1.0 QALY), while the parents were assumed to benefit from an additional 0.07 QALY each as a result of the birth of their child based on the utility gained as a result of fertility as reported for the Health Utilities Index Mark 2.22
Results
Study population
This was a follow-up study of a previously published cohort of infants with suspected monogenic disorders. Of the 80 study participants, 48 were diagnosed: 47 using WES and 1 with conventional testing (not WES). Of the 48 diagnoses, 4 were made on WES data reanalysis (2 after 6 months (ECHS1 and NALCN), 1 after 12 months (KIAA0556), and 1 after 18 months (KAT6A). The diagnosed group included seven deceased infants, three of whom died before a result was available and four of whom died following result disclosures. One diagnosed patient was lost to follow-up. In 44 diagnosed infants, further outcomes data, such as tertiary pediatric services use, were collected after disclosure of the result.
The undiagnosed group comprised 32 infants, of whom 3 were deceased (2 before disclosure of the result and 1 afterwards). One infant was lost to follow-up. In 29 undiagnosed infants, further outcomes data, such as tertiary pediatric services use, were collected after disclosure of the result.
Data such as survival and family reproductive outcomes were collected for the entire cohort (n = 80). The median duration of follow-up after disclosure of the result was 473 days (interquartile range: 411–650). The costs of investigations, medications, procedures, and patient encounters are presented in Supplementary Table 1.
Patient outcomes
Clinical trajectory of undiagnosed infants
Although they initially presented with features suspicious of a monogenic condition, seven undiagnosed infants were judged clinically during the follow-up period to not have a monogenic condition. Symptoms resolved in two infants with uninformative WES results, one presenting with short long bones in the newborn period and the other with severe hepatitis. Postmortem examination identified viral myocarditis in another infant who presented with severe metabolic acidosis. Three infants with congenital abnormalities developed normally and are now considered to have a non-monogenic condition (one field defect and two oculo-auriculo-vertebral spectra). Finally, one infant presenting with abnormal metabolites in the newborn period had a single pathogenic variant identified and confirmed in ETFDH. The metabolic profile and sequencing results together suggested carrier state for glutaric aciduria. Treatment was considered unnecessary and stopped, with the infant discharged from tertiary pediatric care. The annual cost saving from this change in management is estimated to be AU$426.75 (comprising metabolic specialist appointments (three per year; AU$372) and riboflavin supplementation (50 mg three times a day; AU$54.75)).
Clinicians continued to assess and investigate the undiagnosed infants with an ongoing suspicion of a monogenic disorder (n = 29, including live (n = 27) and deceased participants (n = 2)) in parallel with WES data reanalysis. Nine infants (9 out of 29; 31%) underwent further diagnostic investigation (including one Sanger sequencing test and two next-generation sequencing panels) at a cost of AU$15,584.94, with no additional diagnoses made.
Cost-effectiveness of WES data reanalysis
The cost of reanalysis using model 1 (single reanalysis 18 months after the initial result) was AU$11,350 and the cost per diagnosis was AU$2837.50 (95% CI: AU$1568.75 to $10,450.00). Model 1 demonstrated an incremental cost saving of AU$1058.74 (95% CI: –AU$10,501.70 to $1937.17) per additional diagnosis compared with the standard care pathway (Table 1). This was a more cost-effective approach than model 2, which entailed performing two cycles of reanalysis at 6 month intervals over 18 months, with two diagnoses made in the first cycle and one on each subsequent cycle. This service delivery model would have cost AU$29,900.00 in total for this cohort and resulted in a cost per diagnosis of AU$7475.00 (95% CI: AU$3625.00 to $30,400.00). For model 2, the incremental cost per additional diagnosis was estimated to be AU$3578.77 (95% CI: AU$231.71 to $17,003.02) compared with ongoing standard diagnostic investigation (Table 1).
These two models were compared with ongoing standard diagnostic investigation using the cost-effectiveness planes shown in Fig. 1. Each point represents the result of each bootstrapped simulation. The cost-effectiveness planes demonstrate that when compared with the standard diagnostic investigation, model 1 is dominant (i.e., it incurs less cost with a higher number of diagnoses) for a greater number of simulations than model 2 (72.3% of 1000 simulations for model 1 compared with only 3.2% for model 2).
Each point in the scatterplot represents the result of each of 1000 bootstrapped simulations. Points below the horizontal dashed line indicate simulations where WES reanalysis is less costly while providing a higher number of diagnoses compared with standard diagnostic care. This means that for each of these simulations, WES reanalysis is “dominant” (i.e., it costs less while providing greater benefit than standard diagnostic care and is thus considered cost-effective). Points above the horizontal dashed line indicate simulations where the use of WES reanalysis is more expensive but provides a higher number of diagnoses compared with standard diagnostic care. In this situation, WES reanalysis may still be regarded as cost-effective if the incremental cost for one additional diagnosis is less than the cost we are willing to pay for each additional diagnosis
Clinical trajectory of diagnosed infants
No further diagnostic investigations were performed in the diagnosed group. Sixteen study participants had a change in management as a result of WES diagnosis. Of these, four had a change in clinical outcome and/or hospital service use. The costs associated with these changes in management and health outcomes are presented in Table 2. The changes in management that have not so far impacted on clinical outcomes and their costs are presented in Supplementary Table 2. There was little difference in the utilization of tertiary pediatric hospital services between the diagnosed and undiagnosed groups pre- or post-result (Supplementary Table 3).
Family outcomes
Cascade testing
Cascade testing was offered to all parents to clarify the reproductive risks, as well as to siblings where clinically indicated. Of 88 eligible first-degree relatives, 79 were tested for 52 variants at a cost of AU$28,000.
Costs and impact of changes in management: first-degree relatives
Relatives of the infants diagnosed by WES (n = 12) received a molecular diagnosis following cascade testing. Two of these relatives had a change in management as a result. One asymptomatic sibling was diagnosed with thiamine transporter dysfunction syndrome, and commenced on thiamine supplementation at a cost of AU$146 per year ($0.40 daily). She has not experienced any episodes of deterioration from normal functioning (QALY gain of 0.4). A parent was diagnosed with Holt–Oram syndrome and had cardiac surveillance initiated at a cost of AU$329 per year without any cardiac disease detected to date.
Reproductive outcomes
A total of 16 couples sought advice from reproductive genetic services: 2 with undiagnosed children and 14 with diagnosed children. The utilization of reproductive genetic services and reproductive outcomes are summarized in Fig. 2. Further details are available in Supplementary Fig. 1.
In brief, three couples with diagnosed children were seen at a pre-implantation genetic diagnosis (PGD) service. Two decided to proceed with PGD and one was successful in establishing a pregnancy. The third couple decided not to proceed with PGD and were seen by a pre-natal genetics service, together with another 11 couples with diagnosed children. One couple miscarried before pre-natal diagnosis (PND). Four couples proceeded with PND, three of whom were at a 25% recurrence risk, and one at risk of gonadal mosaicism (<1%). One pregnancy was found to be affected on testing. The family continued the pregnancy as the condition is potentially curable by bone marrow transplantation. Another four couples (all at gonadal mosaicism risk; <1%) proceeded with pregnancy without pre-natal testing; one of whom had two successful pregnancies. The total number of ongoing pregnancies in the diagnosed group was nine.
Two families with children who remain undiagnosed have had a pregnancy. Both were seen by a clinical geneticist at a pre-natal genetics service. One terminated the pregnancy at 10 weeks' gestation due to uncertainty regarding the recurrence risk.
The cost of PGD use by this group of families was AU$29,804, of which AU$12,409 was funded by the Australian Government through Medicare, and the remainder by the families. The cost of PND was AU$27,100.37.
Cost–utility analysis of the change in patient management and reproductive planning
Cost–utility analysis accounting for changed management (patient only)
Our analysis demonstrates that genomic testing followed by changes in management as a result of WES diagnosis resulted in a cost saving of AU$1577.88 (95% CI: –AU$205,449.67 to $19,780.25) per additional QALY gained over standard diagnostic care (Table 3).
Cost–utility analysis account for changed management, cascade testing, and reproductive planning in patients and first-degree relatives
Accounting for cascade testing, reproductive service utilization, and outcomes in first-degree relatives, in addition to the impact of altered management in the patients resulted in an incremental cost of AU$8118.70 (95% CI: AU$1961.84 to $38,943.90) per QALY gained (Table 3).
Two cost-effectiveness planes of genomic testing resulting in diagnosis are presented in Supplementary Fig. 2: one considering only the change in clinical management as a result of diagnosis and the other considering all health benefits of having a diagnosis including changes in management, cascade testing, and reproductive outcomes. When only the change in clinical management was considered, achieving a diagnosis as a result of genomic testing was dominant (less cost and an increase in QALY) in 48.5% of the simulations. After accounting for cascade testing and reproductive planning/outcomes in addition to the impact of altered management as a result of genomic testing, 97.8% of the simulation results fall into the top-right quadrant, which suggests that there is greater additional cost per QALY gained when these costs are considered.
Discussion
Genetic conditions are the leading cause of death in infants, and children with genetic conditions are typically high users of tertiary pediatric care services. Genomic testing is a transformative technology that is expected to have a major effect on tertiary pediatric care. However, evidence is necessary to support the case to funders for its implementation, and for services to guide clinical and laboratory decision-making. Our study provides evidence of the broader long-term impacts of this technology on a range of clinical and health economic parameters. In addition, we provide evidence to support policy development and service provision in an area of highly variable clinical and laboratory practice: the reanalysis of stored genomic data.
For our diagnosed patients, the promise of personalized care as a result of genomic sequencing was realized for a large proportion of the cohort. Importantly, the treatments and surveillance initiated were generally of a modest cost and did not result in increased hospital service use. Overall, the changes in clinical management resulting from genomic sequencing across the entire cohort of 80 were found to result in a cost saving of $1578 per QALY gained at one-year follow-up.
Unexpectedly, we found that uninformative WES results had value in supporting the diagnosis of non-Mendelian conditions in 7 patients (8.75% of the cohort). The uninformative genomic sequencing result, combined with the clinical course, enabled an alternate, non-genetic diagnosis to be made, releasing the child from further investigations and unnecessary treatments. Early genomic sequencing offers the greatest opportunities to influence outcomes and reduce the costs of other investigations by making timely diagnoses.1,2,8,10 One of the challenges of initiating genomic testing early in the diagnostic trajectory is that clinical presentations are often poorly differentiated and incomplete, making it difficult to distinguish genetic from non-genetic etiologies. Performing genomic testing in a small number of patients whose subsequent course suggests a non-monogenic etiology may be an inherent risk of early test initiation, but the uninformative results can still be beneficial in contributing to diagnostic certainty, albeit for non-monogenic conditions.
We found that reanalysis of existing WES data was a highly effective diagnostic strategy in patients with uninformative results but ongoing suspicion of a monogenic condition. The diagnostic yield we obtained (14%) is consistent with 3 recent studies totaling over 1000 reanalyzed cases, reporting additional diagnostic yields ranging between 10 and 21% 20 to 36 months after initial analysis.13,14,15 The majority of reanalysis diagnoses have been due to the identification of variants in genes associated with disease following the initial WES analysis. The value of genomic data reanalysis is increasingly acknowledged, but a major challenge for implementation is the development of sustainable models for the laboratory and clinical delivery of this labor-intensive service. The coupling of reanalysis with follow-up clinical genetics appointments at intervals of one to two years provides the opportunity to re-examine the genomic data not only in the context of evolving genetics knowledge, but also in the context of evolving patient phenotype information and family priorities. This model was more cost-effective than undertaking reanalysis every 6 months, with a cost saving of AU$1058 per additional diagnosis compared with usual diagnostic care.
For the infants’ families, the major impact of early genomic diagnosis observed in this cohort is the restoration of parental reproductive confidence. As the study participants underwent WES in the first two years of life, all parents were of reproductive age. A molecular diagnosis in the child was strongly associated with increased use of reproductive genetic services and with subsequent pregnancy, with eight more ongoing pregnancies in the families where the child had a diagnosis compared with those without. All pregnant couples at increased risk of recurrence utilized either PGD or PND, with one couple continuing an affected pregnancy because treatment for the condition was available. The only termination of pregnancy occurred in the “undiagnosed” group, based on uncertainty regarding the recurrence risk. This pattern reflects previous observations from the 1980s of couples postponing and avoiding having more children when PND for a genetic condition was not available.23 Including the cost of cascade testing and reproductive services use resulted in an additional cost of AU$8118 per QALY gained in this cohort over the follow-up period. However, the lifetime value of the additional eight children to families, communities, and Australian society is expected to far exceed the cost of providing reproductive genetic services, and indeed the cost of providing genomic sequencing in the cohort.
This study is limited by its small size, and bootstrapping was conducted to provide an estimate of certainty. The results are not generalizable to patients undergoing genomic testing for different indications or using different service delivery models, as diagnostic yields and the clinical impact of results are likely to be different.24 Collection of clinical utility and cost-effectiveness data in larger clinically ascertained cohorts followed up over extended periods of time will be needed to clarify the lifetime impacts of genomic testing. As clinically ascertained cohorts tend to be small, we encourage standardized data collection around clinical utility parameters and health-care costs across diverse health-care settings to facilitate data analysis and comparison.
Our work provides evidence of the longer-term outcomes of genomic sequencing for patients, their families, and health-care costs. This follow-up study demonstrates that this transformative technology results in improved patient management without a major increase in health-care costs, and in restoration of reproductive confidence in families. The results further strengthen the case for the early use of genomic testing in the diagnostic trajectory. In addition, we show that reanalysis at 18 months—around the time of clinical review—is a cost-effective model for the storage and re-examination of genomic data in clinical service delivery. As health systems worldwide consider the implementation of genomic sequencing in routine clinical practice, these results provide valuable evidence to guide funders and services in the cost-effective use of this technology.
Change history
29 August 2018
The original PDF version of this Article omitted to list Clara L Gaff as a corresponding author and the affiliations were incorrectly labelled as Present Addresses. Furthermore, Tables 1 and 2 have been updated to clarify that the Australian dollar is used for the values. These errors have now been corrected in the PDF and HTML versions of the Article.
References
Stark Z, Tan TY, Chong B, et al. A prospective evaluation of whole-exome sequencing as a first-tier molecular test in infants with suspected monogenic disorders. Genet Med. 2016;18:1090–6.
Willig LK, Petrikin JE, Smith LD, et al. Whole-genome sequencing for identification of Mendelian disorders in critically ill infants: a retrospective analysis of diagnostic and clinical findings. Lancet Resp Med. 2015;3:377–87.
Stark Z, Schofield D, Alam K, et al. Prospective comparison of the cost-effectiveness of clinical whole-exome sequencing with that of usual care overwhelmingly supports early use and reimbursement. Genet Med. 2017;19:867–74.
Tan TY, Dillon OJ, Stark Z, et al. Diagnostic impact and cost-effectiveness of whole-exome sequencing for ambulant children with suspected monogenic conditions. JAMA Pediatr. 2017;171:855–62.
Vissers LE, van Nimwegen KJ, Schieving JH, et al. A clinical utility study of exome sequencing versus conventional genetic testing in pediatric neurology. Genet Med. 2017;19:1055–63.
Griffin BH, Chitty LS, Bitner-Glindzicz M. The 100 000 Genomes Project: what it means for paediatrics. Arch Dis Child Educ Pract Ed. 2016;102:105–7.
Baldridge D, Heeley J, Vineyard M, et al. The Exome Clinic and the role of medical genetics expertise in the interpretation of exome sequencing results. Genet Med. 2017;19:1040–8.
Meng L, Pammi M, Saronwala A, et al. Use of exome sequencing for infants in intensive care units: ascertainment of severe single-gene disorders and effect on medical management. JAMA Pediatr. 2017;171:e173438.
Sawyer SL, Hartley T, Dyment DA, et al. Utility of whole-exome sequencing for those near the end of the diagnostic odyssey: time to address gaps in care. Clin Genet. 2015;89:275–84.
Van Diemen CC, Kerstjens-Frederikse WS, Bergman KA, et al. Rapid targeted genomics in critically ill newborns. Pediatrics. 2017;140:e20162854.
Hayeems RZ, Bhawra J, Tsiplova K, et al. Care and cost consequences of pediatric whole genome sequencing compared to chromosome microarray. Eur J Hum Genet. 2017;25:1303–12.
Bowdin S, Gilbert A, Bedoukian E, et al. Recommendations for the integration of genomics into clinical practice. Genet Med. 2016;18:1075–84.
Wenger AM, Guturu H, Bernstein JA, Bejerano G. Systematic reanalysis of clinical exome data yields additional diagnoses: implications for providers. Genet Med. 2016;19:209–14.
Nambot S, Thevenon J, Kuentz P, et al. Clinical whole-exome sequencing for the diagnosis of rare disorders with congenital anomalies and/or intellectual disability: substantial interest of prospective annual reanalysis. Genet Med. https://doi.org/10.1038/gim.2017.162.
Wright CF, McRae JF, Clayton S, et al. Making new genetic diagnoses with old data: iterative reanalysis and reporting from genome-wide data in 1,133 families with developmental disorders. Genet Med. https://doi.org/10.1038/gim.2017.246.
Jamal SM, Yu JH, Chong JX, et al. Practices and policies of clinical exome sequencing providers: analysis and implications. Am J Med Genet Part A. 2013;161A:935–50.
Gaff CL, M Winship I, Forrest SM, Hansen DP, Clark J, Waring MP et al. Preparing for genomic medicine: a real world demonstration of health system change. Genom Med. 2017;2:16
Monroe GR, Frederix GW, Savelberg SM, et al. Effectiveness of whole-exome sequencing and costs of the traditional diagnostic trajectory in children with intellectual disability. Genet Med. 2016;18:949–56.
Walsh M, Bell KM, Chong B, et al. Diagnostic and cost utility of whole exome sequencing in peripheral neuropathy. Ann Clin Transl Neurol. 2017;4:318–25.
Schofield D, Alam K, Douglas L et al. Cost-effectiveness of massively parallel sequencing for diagnosis of paediatric muscle diseases. NPJ Genom. Med. 2017;2:4
Carroll AE, Downs SM. Improving decision analyses: parent preferences (utility values) for pediatric health outcomes. J Pediatr. 2009;155:21–5.
Scotland GS, McLernon D, Kurinczuk JJ, et al. Minimising twins in in vitro fertilisation: a modelling study assessing the costs, consequences and cost–utility of elective single versus double embryo transfer over a 20-year time horizon. BJOG. 2011;118:1073–83.
Evers-Kiebooms G, Denayer L, Van, den Berghe H. A child with cystic fibrosis: II. Subsequent family planning decisions, reproduction and use of prenatal diagnosis. Clin Genet. 1990;37:207–15.
Dragojlovic N, Elliott AM, Adam S, et al. The cost and diagnostic yield of exome sequencing for children with suspected genetic disorders: a benchmarking study. Genet Med. https://doi.org/10.1038/gim.2017.226.
Acknowledgements
We thank all collaborators in the Melbourne Genomics Health Alliance Demonstration Project, and K. Hood for administrative support with preparation of the manuscript.
Funding
The study was funded by the founding organizations of the Melbourne Genomics Health Alliance (Royal Melbourne Hospital, Royal Children’s Hospital, University of Melbourne, Walter and Eliza Hall Institute, Murdoch Children's Research Institute, Australian Genome Research Facility, and CSIRO) and the State Government of Victoria (Department of Health and Human Services). The involvement of AGRF was supported by sponsorship from Bioplatforms Australia and the NCRIS program.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure
The authors declare no conflicts of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Stark, Z., Schofield, D., Martyn, M. et al. Does genomic sequencing early in the diagnostic trajectory make a difference? A follow-up study of clinical outcomes and cost-effectiveness. Genet Med 21, 173–180 (2019). https://doi.org/10.1038/s41436-018-0006-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-018-0006-8
Keywords
This article is cited by
-
Systematic reanalysis of genomic data by diagnostic laboratories: a scoping review of ethical, economic, legal and (psycho)social implications
European Journal of Human Genetics (2024)
-
Decoding complex inherited phenotypes in rare disorders: the DECIPHERD initiative for rare undiagnosed diseases in Chile
European Journal of Human Genetics (2024)
-
Rare diseases, common barriers: disparities in pediatric clinical genetics outcomes
Pediatric Research (2023)
-
Reanalysis of whole-exome sequencing (WES) data of children with neurodevelopmental disorders in a standard patient care context
European Journal of Pediatrics (2023)
-
Bayesian cost-effectiveness analysis of Whole genome sequencing versus Whole exome sequencing in a pediatric population with suspected genetic disorders
The European Journal of Health Economics (2023)