Abstract
Purpose
Integration of gene panels in the diagnosis of hereditary breast and ovarian cancer (HBOC) requires a careful evaluation of the risk associated with pathogenic or likely pathogenic variants (PVs) detected in each gene. Here we analyzed 34 genes in 5131 suspected HBOC index cases by next-generation sequencing.
Methods
Using the Exome Aggregation Consortium data sets plus 571 individuals from the French Exome Project, we simulated the probability that an individual from the Exome Aggregation Consortium carries a PV and compared it to the estimated frequency within the HBOC population.
Results
Odds ratio conferred by PVs within BRCA1, BRCA2, PALB2, RAD51C, RAD51D, ATM, BRIP1, CHEK2, and MSH6 were estimated at 13.22 [10.01–17.22], 8.61 [6.78–10.82], 8.22 [4.91–13.05], 4.54 [2.55–7.48], 5.23 [1.46–13.17], 3.20 [2.14–4.53], 2.49 [1.42–3.97], 1.67 [1.18–2.27], and 2.50 [1.12–4.67], respectively. PVs within RAD51C, RAD51D, and BRIP1 were associated with ovarian cancer family history (OR = 11.36 [5.78–19.59], 12.44 [2.94–33.30] and 3.82 [1.66–7.11]). PALB2 PVs were associated with bilateral breast cancer (OR = 16.17 [5.48–34.10]) and BARD1 PVs with triple-negative breast cancer (OR = 11.27 [3.37–25.01]). Burden tests performed in both patients and the French Exome Project population confirmed the association of PVs of BRCA1, BRCA2, PALB2, and RAD51C with HBOC.
Conclusion
Our results validate the integration of PALB2, RAD51C, and RAD51D in the diagnosis of HBOC and suggest that the other genes are involved in an oligogenic determinism.
Similar content being viewed by others
INTRODUCTION
The development of high-throughput sequencing (or next-generation sequencing, NGS) has created an unprecedented revolution in the molecular diagnosis of inherited diseases. In this context, hereditary breast and ovarian cancer syndrome (HBOC) represents an interesting model. Indeed, molecular diagnosis of HBOC was initially restricted to the identification of germline pathogenic variants (PVs) within the two main genes, BRCA1 and BRCA2.1,2 Today, many medical laboratories have integrated bioinformatics expertise quality and insurance (i.e., ISO 15189) in their practice, which are all required for NGS. These laboratories are performing molecular diagnosis of HBOC by NGS analysis of gene panels.3,4,5,6 We and other teams had previously reported that in families with suspected HBOC, almost 50% of PVs were identified in genes other than BRCA1 and BRCA2.4 The involvement of non-BRCA genes in breast cancers has been clearly established, including TP53, whose germline PVs cause Li-Fraumeni syndrome; TP53 germline PVs are the main cause of very early breast cancers, occurring before 31 years of age.7 In contrast, germline PVs within other genes and the associated levels of cancer risk induced by these PVs are a matter of debate and the use of gene panels in HBOC diagnosis is a subject of controversy in medical laboratories.8, 9 Indeed, integration of gene panels in the diagnosis of HBOC requires a careful evaluation of the risk associated with the genetic variants detected in each gene.
For some non-BRCA putative HBOC genes, progressive accumulation of segregation, statistical, and functional data knowledge has refined the risk evaluation.10 For instance, PVs in PALB2 have clearly been shown to be associated with an increased risk of breast cancer, but according to the publications, the level of risk varies from moderate to a high risk that is of the same magnitude as the risk conferred by BRCA2 PVs.11,12,13 Consequently, the appropriate clinical management of PALB2 PVs carriers is still debated. In the same manner, the levels of ovarian cancer risk associated with RAD51C and RAD51D PVs appear to be sufficient to propose prophylactic salpingo-oophorectomy after 50 years of age, although the estimated risk confidence intervals are large and cancer consultants hesitate to systematically recommend risk-reducing surgery.14, 15 In contrast, the estimated risks associated with germline PVs detected in other genes commonly integrated in gene panel diagnostics for HBOC, such as BRIP1 for ovarian risk,16 or BARD117 and FANCM18 for breast cancer risk, are either contradictory or too low to use them in order to guide clinical management and genetic counseling within the families.
This imprecision of the estimated levels of risks is, in part, explained by the low incidence of PVs detected in these genes. Consequently, thousands of cases and appropriate controls are required to document significant enrichment of PVs. A very large association study, performed on 41,611 breast cancer patients and Exome Aggregation Consortium (ExAC) reference-appropriate controls, recently reported that besides the BRCA, CDH1, PTEN, and TP53 genes only pathogenic variants in PALB2 were associated with a high breast-cancer risk (odds ratio (OR), 7.46). Variants within ATM, BARD1, CHEK2, and RAD51D were found to be associated with moderately increased risks with ORs estimated at 2.78, 2.16, 1.48, and 3.07, respectively.13 Although that study provided useful information for diagnostic laboratories, it might be exposed to the risk of the genetic heterogeneity of the analyzed samples. These case-control studies must take into account the stratification bias induced by the geographical ancestral origin of the controls13, 19, 20 and they should also consider the potential bias resulting from the allelic frequency provided by the ExAC dataset. Indeed, allelic frequencies in the ExAC database are deduced from the adjusted counts obtained after quality-control filtering of the sequenced alleles. Comparison between these allelic frequencies in controls and patients, without taking into account the heterogeneity of this measurement, may impact the risk estimation.
Here we developed a model that allows calculation of the probability that a case or a control is a carrier of a variant in order to analyze the association with HBOC of variants detected in 34 genes among 5131 French patients. To limit the bias and provide accurate estimates of the risks associated with the detected variants, we restricted the analysis to patients of French origin and we also performed burden tests using individual genomic data from 571 French control genotypes sequenced by the French Exome (FREX) Consortium.
MATERIALS AND METHODS
Patients and controls
Patients analyzed in this study have been seen in the context of genetic counseling and all fulfilled at least one of the criteria presented in Fig. 1. 5,131 French patients were studied. All patients gave informed consent for genetic analysis. We collected genomic data from 571 control individuals from six French regions sequenced by the FREX project (www.france-genomique.org/spip/spip.php?article158). All individuals were tested for their geographic origin, using multiple correspondence analysis of common single-nucleotide polymorphisms within 34 HBOC genes (Supplementary Table S1), with a minor allele frequency >5%, as determined by the 1000 Genomes Project (phase 3). After filtering, 281 variants were thus included. In order to compute posterior probabilities of the individual’s geographical origin, a linear discriminant model was adjusted on the first relevant axis from the multiple correspondence analysis. Individuals with a posterior probability of non-European origin >95% were excluded from analyses (arbitrary threshold).
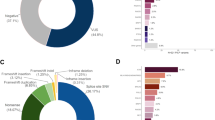
Clinical presentation of the patients analyzed in this study and relative distribution of PVs detected with NGS. a Overview of the individual and familial presentation of the patients included in the analysis; a patient classification can revolve around several criteria. b Percentages in the relative distribution of PVs detected were based on the number of times the gene was sequenced, depending on the version of the capture design. NGS, next-generation sequencing; PV, pathogenic variant
Sample preparation, sequencing, and bioinformatic analyses
DNA was extracted from peripheral blood, using the Agencourt Genfind V2 on Biomek FX (Beckman, Villepinte, France). DNA was sonicated using a Covaris S2 (Covaris, MS. Samples were prepared with SPRIworks HT (Beckman). Illumina adapters were replaced by indexed adapters (NEB, Milan, Italy) previously published by Huentelman’s team.21 We designed two different Sureselect libraries (Agilent, Santa Clara, CA), covering. variable number of HBOC genes (Supplementary Table S1). The enrichment process was performed according to Kenny et al.4, 22 The protocol was robotized on a Biomek FX. Libraries were sequenced on a Miseq or NextSeq500 (Illumina, San Diego, CA). Data from patients were generated with CASAVA v.1.8 suite from Illumina. Variants from the 5131 variant call formats generated by CASAVA were excluded if depth <20 or a quality score (QUAL) <100 for single-nucleotide variants or QUAL <500 for indels, ensuring sensitivity and specificity, as previously described.4 For one gene, individual data from the 5131 patients were excluded if at least one exonic position did not reach 20× coverage. Also if more than 20% of the patients were not covered with at least 20× on the exons, these specific regions were excluded from the final analysis. This corresponded to <1% of the targeted region (i.e. five exons). FREX genomic data were generated using Agilent SureSelect Human All Exon kits. Final libraries were sequenced on a HiSeq2000 or 250020 (Illumina). Individual genome variant call formats were generated by the FREX consortium. Best practices as edited by the Broad Institute were used to determine the genotypes. Variants from FREX with <5% of missing data were kept when VQSR values reached 99.5% for single-nucleotide variants and 99% for indels. Only variants with a mutated allele fraction above 25% were kept. We validated that CASAVA was not more sensitive than the best practices as edited by the Broad Institute in detecting variants, ensuring that the following analysis did not overestimate association calculations by favoring the detection of pathogenic variants in the control population (data not shown).
Probability calculation and statistical analyses
Genomic regions of interest were defined as where the exome and HBOC gene panels intersected (Supplementary Table S1) and we included only variants in these regions in our study. The adjusted allele count provided by ExAC for the non-Finnish European (NFE) population in the ExAC non-TCGA subset data (ExAC.r0.3.nonTCGA.sites.vep.vcf.gz) and corresponding to individuals with genotype quality ≥20 and depth ≥10 was used. Pathogenic ExAC variants classified in the “non-pass” category, as defined below, in the variant call format, were checked manually.
All the variants detected in the patients or in the ExAC and the FREX databases were annotated using ANNOVAR (v2015-12-14 database 2014-10) and Alamut-BATCH (v1.4.2 database 1.4.-2015.11.02 GRCH37; IBS, Rouen, France). Annotated variants were categorized using the standard ACMG criteria.23 Analyses were restricted to variants with a minor allele frequency <1% in all three populations and considered here in the PM2 category (low allele frequency). For genes with alternative transcripts, we considered the transcript with the most pathogenic prediction. Pathogenic or likely pathogenic variants were called as PVs if any of the following applied:
- 1.
The corresponding clinical significance of variants detected was based on consensus data integrated in the French UMD-BRCA1/BRCA2 databases, the IARC TP53 Database or the Insight database for mismatch repair genes (PP5 category)
- 2.
They introduced a premature termination codon (PVS1 category)
- 3.
They affect the canonical AG/GT splice sites (PVS1 category)
- 4.
MaxEntScan and SpliceSiteFinder scores predicted a complete abolition of the donor or the acceptor splice site even if a value was missing with a very high level of confidence, i.e., “GC site” not predicted by MaxEntScan” (strictly PP3 category, but, this very high evidence of pathogenicity led us to categorize them as PVs)24
- 5.
They abolished the translation initiation codon (PVS1 category).
For the other variants, we considered only variants in the PP3 category with a high level of confidence in their pathogenicity, called VUS (variants of uncertain significance). We took VUS corresponding either to
- a.
missense variants predicted as pathogenic according to Mutation Taster, SIFT, and Polyphen-2 (Hum-Div) algorithms, or
- b.
variants outside the canonical AG/GT splice sites but predicted to alter splicing (50% decrease according to MaxEntScan and SpliceSiteFinder scores, or a complete abolition with one of these algorithms and a decrease with the other.
These selected variants, if they are more prevalent in HBOC families (PS4 category) are considered as likely pathogenic or pathogenic, according to ACMG rules. Alternatively to these prioritization rules, we used the Combined Annotation Dependent Depletion (CADD) algorithm25 to select variants with a minor allele frequency <1%.
We evaluated the enrichment in PVs and/or VUS within 34 HBOC genes. Ppatient, PFREX, and PExAC are defined as the probabilities that an individual, respectively from the 5,131 patients, FREX, and ExAC (NFE) population samples, carries a pathogenic variation. Ppatient and PFREX were estimated as proportions with an usual Casagrande-Pike confidence interval. PExAC is equal to {1 – the probability of an ExAC individual to not carry a pathogenic variation}. Assuming the independence of these rare pathogenic variations, i.e., no linkage disequilibrium, PExAC can be formulated as \(\left\{ {1- \mathop {\prod }\limits_{i= 1}^k 1- \frac{{{\mathrm{AC}}\_{\mathrm{NFE}}_{\mathrm{i}}- {\mathrm{Hom}}\_{\mathrm{NFE}}_i}}{{({\mathrm{AN}}\_{\mathrm{NFE}}_i{\mathrm{/}}2)}}} \right\}\), where \(\frac{{{\mathrm{AC}}\_{\mathrm{NFE}}_i- {\mathrm{Hom}}\_{\mathrm{NFE}}_i}}{{({\mathrm{AN}}\_{\mathrm{NFE}}_i{\mathrm{/}}2)}}\) is the estimated probability that an individual from the ExAC NFE population carries the ith variation between the k pathogenic variants identified in each analyzed gene. With ExAC notation, AN_NFEi, AC_NFEi and Hom_NFEi are, respectively, the number of alleles considered for the adjusted count, the number of alternative alleles, and the number of homozygous individuals counted in the ExAC NFE population for this ith variant. The interval of confidence of PExAC was empirically determined by modeling each probability \(\frac{{{\mathrm{AC}}\_{\mathrm{NFE}}_i- {\mathrm{Hom}}\_{\mathrm{NFE}}_i}}{{({\mathrm{AN}}\_{\mathrm{NFE}}_i{\mathrm{/}}2)}}\) using a Beta distribution with (AC_NFEi−Hom_NFEi; AN_NFEi/2−(AC_NFEi, and Hom_NFEi)) as parameters.26 In the same manner, the Patient/ExAC OR was empirically determined by jointly modeling PExAC and Ppatient and calculating \(\frac{{p_{{\mathrm{patient}}}.(1- p_{{\mathrm{ExAC}}})}}{{\left( {1- p_{{\mathrm{patient}}}} \right).p_{{\mathrm{ExAC}}}}}\) for each simulation. 100,000 simulations were performed for each estimate. The Patient/FREX OR and its confidence interval were estimated conventionally. Two complementary burden tests, corresponding to the cohort allelic sum test (CAST) and weighted-sum test (WST), were used to test the association between HBOC and pathogenic variants, using the FREX population as the reference.
RESULTS
From 5,131 patients with suspected HBOC, 270 were excluded because the quality of sequencing did not fulfill quality criteria and 160 others were excluded because they were either related to another index case or because clinical criteria did not meet the prerequisites for the statistical study (Supplementary figure S1). The multiple correspondence analysis revealed a superposition of the projections of the genotypes from analyzed patients and FREX controls (Fig. 2). The analysis also detected and excluded from the following analysis 292 additional patients and 12 FREX individuals that are probably not of European origin, according to the 1000 Genomes training dataset. In the 4,409 remaining patients, the frequencies of PVs in BRCA2, BRCA1, CHEK2, ATM, PALB2, RAD51C, TP53, and BRIP1 were 3.9%, 3.7%, 1.1%, 1.0%, 0.9%, 0.5%, 0.5%, and 0.5%, respectively (Supplementary table S1 and Fig. 1). Fewer than 0.3% of the patients had PVs in MSH2, MSH6, MLH1, or PMS2. The incidence of MUTYH PVs was 2.4%, consistent with the documented frequency of recurrent PVs in the French population.27 This high incidence would probably confuse the results and so the MUTYH gene was removed from the subsequent analyses. Almost half (48.6%) of our detected PVs were in genes other than BRCA1 or BRCA2, confirming the genetic heterogeneity of HBOC. Half of the VUS were in just six genes: ATM, MLH1, BRIP1, NBN, CHEK2, and FAM175A (Supplementary Figure S2). Patients with PVs had an earlier mean age of breast-cancer diagnosis than non-carrier patients within the same gene, not only for BRCA1 (41.8 years, p < 0.001), or for BRCA2 (44.6 years, p < 0.05) but also for CHEK2 (42.2 years, p < 0.01) and ATM (42.5 years, p < 0.05) (Supplementary table S2).
Multiple correspondence analysis and selection of patients and FREX individuals based on ancestral geographic origin. Projection of 1,000 genomes, patients, and FREX individual data, respectively on the first discriminant plane for scatter plots A1–3 and on the second and third axes-plane for scatter plots B1–3. Discriminant axes were fitted on the multiple correspondence analyses of the 1000 genomes training set. Patients and FREX individuals (gray dots) were predicted to be of non-European origin according to this model (p > 95%) and thus excluded from the analysis. FREX, French Exome Project consortium.
Next we analyzed the association of PVs with HBOC, by determining the OR of the calculated probabilities of being a PV carrier in patients and controls. According to our model, the simulated confidence interval derived from OR provides an estimation of a potential association of PVs with HBOC and indirectly with a risk of cancer. First, to quantify a possible bias related to the simulations, we evaluated the enrichment in rare, likely benign, synonymous variants (minor allele frequency < 1%) that probably do not induce a splice anomaly (i.e., not classified as PVs or VUS). Assuming that these synonymous variants should not increase cancer risk, an association of HBOC and rare synonymous variants for a gene would indicate a putative bias. Synonymous variants were indeed enriched in four genes: ATM (OR = 1.40 [1.17–1.64]), MLH1 (OR = 1.66 [1.08–2.38]), RAD50 (OR = 1.66 [1.25–2.15]), and RAD51 (OR = 2.33 [1.42–3.55]) (Supplementary table S3). Therefore, the ORs derived from subsequent analysis of these four genes will have to be interpreted carefully. We estimated the ORs for PVs within BRCA1 and BRCA2 on the entire HBOC series to be 13.22 [10.01–17.22] and 8.61 [6.78–10.82] respectively (Table 1, Fig. 3), which is close to previous estimates.28 In families with breast cancer only, the BRCA ORs were slightly lower at 9.72 [7.07–13.07] and 8.31 [6.40–0.63] respectively. PVs in BRCA1 were more associated than those in BRCA2 with HBOC families with ovarian cancer (OR = 22.56 [16.08–30.66] vs. OR 9.40 [6.65–12.80]) respectively. In HBOC families with triple-negative breast cancer index case, PVs within BRCA1 were associated with high OR (OR = 36.59 [25.05–51.56]) and PVs within BRCA2 at a lower level (OR = 10.34 [6.43–15.41]). BRCA1 and BRCA2 PVs were also associated with families with a bilateral breast cancer index case (respectively OR = 18.94 [10.66–30.19] and 10.26 [5.74–16.33]) and with early-age-of-diagnosis index cases (respectively OR = 16.05 [11.49–21.87] and 10.72 [7.98–14.08]). We calculated the OR for PALB2 PVs at 8.22 [4.91–13.05] (close to that of BRCA2 PVs). PALB2’s OR was higher in HBOC families with breast cancer only, at 10.25 [6.03–16.40]) but no significant association was found in families with at least one ovarian case. PALB2 PVs were highly associated with families whose index case presented a triple-negative or bilateral breast cancer or an early age at diagnosis (OR = 8.75 [2.63–19.24]; 16.19 [5.48–34.22]; 8.18 [4.00–14.47], respectively). We detected a significant association of RAD51C and RAD51D PVs with HBOC patients (respectively, 4.54 [2.55–7.48] and 5.23 [1.46–13.17]), but this effect was maintained only in families with ovarian cancer (11.36 [5.78–19.59] and 12.44 [2.94–33.30]). BRIP1 PVs were mildly associated with HBOC families with at least one ovarian cancer case (OR 3.82 [1.66–7.11]) and, surprisingly, MRE11A PVs are strongly associated with families with only ovarian cancers 10.57 [2.66–25.15]). PVs in CHEK2 and BRIP1 were also weakly associated with HBOC (OR respectively 1.67 [1.18–2.27] and 2.49 [1.42–3.97]). There was a slight association (OR = 2.50 [1.12–4.67]) of PVs with MSH6 and HBOC, as suggested previously.29 More unexpectedly, PVs in BARD1 were associated only with HBOC families where the index case developed a triple-negative breast cancer (OR = 11.27[3.37–25.01]).
Distribution of odds ratios evaluating the association between HBOC and PVs in the patient population and the ExAC population. Violin plots with Gaussian kernel density estimation representing Patient/ExAC ORs distribution among 100,000 simulations. Only results comparing PVs with a significant OR greater than one are represented. The violin-labeled PVs show the distribution of the Patient/ExAC ORs for PVs; the ones labeled RS stand for the Patient/ExAC ORs for rare synonymous variations as a base enrichment value (validation step). The colored areas of the violin represent the 95% confidence interval of the ORs. The black box represents the first and third quartiles’ interval. The white point gives the average of the simulations. ExAC, Exome Aggregation Consortium; HBOC, hereditary beast and ovarian cancer; OR odds ratio; PV, pathogenic variant
We next performed the same analyses after including the VUS. This extension dramatically reduced the associations or abolished them (Supplementary Table S4). When the analysis was restricted to VUS only (Supplementary table S5), we detected a weak association of VUS with BRCA1, BRCA2, BRIP1, MRE11A (significant OR from 1.58 to 2.27). Even more unexpected was a moderate association between HBOC and VUS in BARD1 (OR = 3.31 [1.93–5.22]).
Next we used CADD to select variants (Supplementary Table S6). We found that variants with scaled C-score >20, association effects had low significant OR and variants with scaled C-score >30 had the highest significant OR. With this last threshold, OR confirmed the association between HBOC and variants in BRCA1 (12.45 [8.51–17.72]), BRCA2 (3.90 [3.01–4.96], ATM (1.56 [1.23–1.94]), BRIP1 (1.85 [1.28–2.55]), PALB2 (5.05 [2.83–8.29], RAD51C (2.70 [1.25–4.94]), and RAD51D (2.60 [1.17–4.84]). Other significant associations were found with BARD1 (2.75 [1.46–4.58]) and TP53 (3.77 [1.12–8.90]).
In order to strengthen previous results data from FREX controls were used to perform CAST and WST burden tests (Table 2). These tests confirmed the associations between HBOC and BRCA1, BRCA2, PALB2, and RAD51C PVs. We could not calculate an OR for PALB2, RAD51C, and RAD51D because of the limited size of our sample and the absence of PVs in the FREX dataset. No other significant association was detected using the FREX data testing PVs. CADD analysis confirmed the association of HBOC with PVs of BRCA1 and BRCA2 (Supplementary Table S7).
DISCUSSION
We show here that it is feasible to carefully reevaluate the risks induced by pathogenic variants of genes sequenced by diagnostic laboratories. This reevaluation is possible using heterogeneous control datasets, including recalibrated data from the ExAC consortium, the 1000 Genomes project, and appropriate controls such as the FREX dataset. We used a stringent analysis workflow based on the control of stratification, on the accuracy of the clinical data, and on the quality of NGS data, to demonstrate an enrichment of PVs in patients. Importantly, we could also replicate the data by performing burden tests using the appropriate FREX Project dataset as control (Table 2). This confirms that it is possible, with these cautions, to evaluate disease risk by comparing allelic frequencies observed in large series of patients to those deduced from massive reference datasets, such as ExAC.13 Based on our results from recruitment of selected families with high genetic risks of cancer, we speculate that breast or ovarian cancer risk might be overestimated for PV carriers, if a familial model is not considered.10 Furthermore, because we excluded variants leading to mosaics, clonal hematopoiesis produces negligible bias into our workflow (in fact, no selected PVs or VUS showed a mutated allelic fraction smaller than 35%). The odds ratios we calculate describe the level of association of PVs with HBOC and the comparison of significant ORs between genes may allow us to estimate the level of cancer risk of PVs of each gene, because some of them are now completely known (i.e., BRCA1 and BRCA2). Prioritization of variants is a crucial step and our classification based on ACMG criteria outperforms the CADD algorithm regarding known levels of cancer risk induced by pathogenic variants in BRCA1 and BRCA2. Nevertheless, CADD brought complementary information and should be adapted to discover association effects from exomes as variants cannot be curated manually, as in small gene panels.
The ORs that we calculated from the PV analysis of this large series of patients validate our integrative approach and place BRCA1 and BRCA2, and firmly now, PALB2, RAD51C, and RAD51D in the molecular diagnosis of HBOC. In contrast, the lower OR values observed for the other genes suggest that they could be involved in an oligogenic determinism of breast and ovarian cancers (Table 1). We confirmed that PALB2 PVs are associated with a high risk of breast cancer as strongly as BRCA2 PVs.11 This level of breast cancer risk justifies the integration of PALB2 into genetic testing of patients referred for HBOC and the use of PALB2 PVs in genetic counseling. This confirms that the clinical management of PALB2 PV carriers should be the same as that recommended for BRCA2 PV carriers according to National Comprehensive Cancer Network guidelines but restricted to breast cancer risk. Indeed, in this series, a high OR was found in breast-cancer-only families considering PVs in PALB2, equivalent to the OR calculated with BRCA2. However, no significant association of PALB2 PVs was found with families with at least one ovarian cancer case. Interestingly, PALB2 PVs were also associated with HBOC families whose index case was triple-negative or bilateral breast cancer and with a younger age of diagnosis (<45). This observation prompts us to prioritize rescreening for PABL2 variants in the index cases, or in a first-degree relative, with one of these three last criteria if BRCA1 or BRCA2 PVs are absent. We also suggest prophylactic salpingo-oophorectomy for menopausal carrier women in ovarian cancer families with RAD51C and RAD51D PVs as ORs conferred by PVs detected within these genes are comparable to those conferred by BRCA2 PVs.15, 30,31,32
We also confirmed the moderate cancer risks induced by CHEK2 and BRIP1 PVs. However, considering the moderate ORs for breast cancer conferred by CHEK2 PVs (1.77 [1.20–2.48]), and for ovarian cancer induced by BRIP1 PVs 3.82 [1.66–7.11], it appears inappropriate to base clinical management on these variants.12, 33, 34 Our results are consistent with previous publication describing the absence of significant risk of breast cancer and moderate risk of ovarian cancer with PV of BRIP1.16, 35, 36 Interestingly, BARD1 PVs were associated with ovarian cancer and bilateral breast cancer (Table 1) as previously demonstrated.16 Overall, then, BARD1 PVs are probably responsible for risk of cancer, and maybe confer high risk, albeit restricted to particular types. Generally, assessment of risk should take into account the precise tumor phenotype. More effort and wider population studies are needed to collect larger data sets to specify risks in subgroups of tumor. Our results suggesting a high ovarian cancer risk in MRE11a PV carriers thus need to be confirmed by other studies. Interestingly, a previous study had suggested modification of the ovarian cancer risk by rare haplotypes of MRE11a.37 Finally, we did not find any association of breast cancer with TP53 variants, or with a lower association effect than expected regarding CADD analysis (Supplementary Table S6), despite the well-established fact that PVs in TP53 confer a high risk of early-onset breast cancer. This apparently unexpected result has a precedent in previous studies which did not detect a significant association either.9, 13 The low frequency of TP53 PVs could explain this result, but we suspect a bias in our recruitment may also be the cause. Indeed early-onset breast cancer cases linked owing to TP53 PVs have a high probability of Li Fraumeni syndrome and these patients were not referred to our laboratory for HBOC, since the Li Fraumeni diagnosis was strongly suspected or had already been established.
We also confirmed the known moderate risk conferred by ATM PVs. Nevertheless, the fact that we detected an association with rare synonymous variants (Supplementary Table S3)12, 38 limits the applicability of this observation. Similarly, we found an effect with synonymous variants of MSH2, RAD50, and RAD51. These effects could be induced by a stratification bias or could be due to a true signal of association underlining a low level of risk. According to the ORs found, PVs in CHEK2, ATM, and BRIP1 and perhaps also FAM175A, MLH3, PMS1 (Table 2) confer mild risks. These results, and other described moderate- or low-risk factors, are not individually useful for care management, but they would have more clinical utility if they were integrated in an oligogenic risk evaluation model.39
The risk evaluations in this study confirm the relevance for HBOC diagnosis of a panel gene-sequencing strategy including not only BRCA1 and BRCA2 but also PALB2, RAD51C, and RAD51D. Based on this study, we consider that the refinement of risk in the “other genes” suspected to be involved in HBOC does not allow their routine analysis in the context of medical laboratories, but it does prefigure the development of oligogenic risk scores, based not only on frequent variants but also including rare pathogenic variants of these genes.
References
Miki Y, Swensen J, Shattuck-Eidens D, et al. strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71.
Wooster R, Bignell G, Lancaster J, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–92.
Buys SS, Sandbach JF, Gammon A, et al. study of over 35,000 women with breast cancer tested with. 25-gene panel of hereditary cancer genes. Cancer. 2017;123:1721–30.
Castéra L, Krieger S, Rousselin A, et al. Next-generation sequencing for the diagnosis of hereditary breast and ovarian cancer using genomic capture targeting multiple candidate genes. Eur. Hum Genet. 2014;22:1305–13.
Shirts BH, Casadei S, Jacobson AL, et al. Improving performance of multigene panels for genomic analysis of cancer predisposition. Genet Med. 2016;18:974–81.
Susswein LR, Marshall ML, Nusbaum R, et al. Pathogenic and likely pathogenic variant prevalence among the first 10,000 patients referred for next-generation cancer panel testing. Genet Med. 2016;18:823–32.
Bougeard G, Renaux-Petel M, Flaman J-M, et al. Revisiting Li-Fraumeni syndrome from TP53 mutation carriers. J Clin Oncol. 2015;33:2345–52.
Li J, Meeks H, Feng B-J, et al. Targeted massively parallel sequencing of. panel of putative breast cancer susceptibility genes in. large cohort of multiple-case breast and ovarian cancer families. J Med Genet. 2016;53:34–42.
Thompson ER, Rowley SM, Li N, et al. Panel testing for familial breast cancer: calibrating the tension between research and clinical care. J Clin Oncol. 2016;34:1455–9.
Easton DF, Pharoah PDP, Antoniou AC, et al. Gene-panel sequencing and the prediction of breast-cancer risk. N Engl. Med. 2015;372:2243–57.
Antoniou AC, Foulkes WD, Tischkowitz M. Breast-cancer risk in families with mutations in PALB2. N Engl. Med. 2014;371:1651–2.
Southey MC, Goldgar DE, Winqvist R, et al. PALB2, CHEK2 and ATM rare variants and cancer risk: data from COGS. J Med Genet. 2016;53:800–11.
Couch FJ, Shimelis H, Hu C, et al. Associations between cancer predisposition testing panel genes and breast cancer. JAMA Oncol. 2017;3:1190–6.
Norquist BM, Harrell MI, Brady MF, et al. Inherited mutations in women with ovarian carcinoma. JAMA Oncol. 2016;2:482–90.
Song H, Dicks E, Ramus SJ, et al. Contribution of germline mutations in the RAD51B, RAD51C, and RAD51D genes to ovarian cancer in the population. J Clin Oncol. 2015;33:2901–7.
Ramus SJ, Song H, Dicks E, et al. Germline mutations in the BRIP1, BARD1, PALB2, and NBN genes in women with ovarian cancer. J Natl Cancer Inst. 2015;107:djv214.
Young EL, Feng BJ, Stark AW, et al. Multigene testing of moderate-risk genes: be mindful of the missense. J Med Genet. 2016;53:366–76.
Neidhardt G, Hauke J, Ramser J, et al. Association between loss-of-function mutations within the FANCM gene and early-onset familial breast cancer. JAMA Oncol. 2016;3:1245–8.
Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91.
Nicolas G, Charbonnier C, Wallon D, et al. SORL1 rare variants:. major risk factor for familial early-onset Alzheimer’s disease. Mol Psychiatry. 2016;21:831–6.
Craig DW, Pearson JV, Szelinger S, et al. Identification of genetic variants using bar-coded multiplexed sequencing. Nat Methods. 2008;5:887–93.
Kenny EM, Cormican P, Gilks WP, et al. Multiplex target enrichment using DNA indexing for ultra-high throughput SNP detection. DNA Res. 2011;18:31–38.
Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants:. joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Houdayer C, Caux-Moncoutier V, Krieger S, et al. Guidelines for splicing analysis in molecular diagnosis derived from. set of 327 combined in silico/in vitro studies on BRCA1 and BRCA2 variants. Hum Mutat. 2012;33:1228–38.
Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–5.
Lee JC, Sabavala DJ. Bayesian estimation and prediction for the beta-binomial model. J Bus Econ Stat. 1987;5:357–67.
Buecher B, Bonaïti C, Buisine M-P, Colas C, Saurin J-C. French experts report on MUTYH-associated polyposis (MAP). Fam Cancer. 2012;11:321–8.
Mavaddat N, Peock S, Frost D, et al. Cancer risks for BRCA1 and BRCA2 mutation carriers: results from prospective analysis of EMBRACE. J Natl Cancer Inst. 2013;105:812–22.
Win AK, Lindor NM, Winship I, et al. Risks of colorectal and other cancers after endometrial cancer for women with Lynch syndrome. J Natl Cancer Inst. 2013;105:274–9.
Loveday C, Turnbull C, Ruark E, et al. Germline RAD51C mutations confer susceptibility to ovarian cancer. Nat Genet. 2012;44:475–6.
Meindl A, Hellebrand H, Wiek C, et al. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as. human cancer susceptibility gene. Nat Genet. 2010;42:410–4.
Sopik V, Akbari MR, Narod SA. Genetic testing for RAD51C mutations: in the clinic and community. Clin Genet. 2015;88:303–12.
Leedom TP, LaDuca H, McFarland R, Li S, Dolinsky JS, Chao EC. Breast cancer risk is similar for CHEK2 founder and non-founder mutation carriers. Cancer Genet. 2016;209:403–7.
Muranen TA, Greco D, Blomqvist C, et al. Genetic modifiers of CHEK2*1100delC-associated breast cancer risk. Genet Med. 2016;19:599–603.
Easton DF, Lesueur F, Decker B, et al. No evidence that protein truncating variants in BRIP1 are associated with breast cancer risk: implications for gene panel testing. J Med Genet. 2016;53:298–309.
Rafnar T, Gudbjartsson DF, Sulem P, et al. Mutations in BRIP1 confer high risk of ovarian cancer. Nat Genet. 2011;43:1104–7.
Rebbeck TR, Mitra N, Domchek SM, et al. Modification of ovarian cancer risk by BRCA1/2-interacting genes in. multicenter cohort of BRCA1/2 mutation carriers. Cancer Res. 2009;69:5801–10.
Renwick A, Thompson D, Seal S, et al. ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nat Genet. 2006;38:873–5.
Shieh Y, Eklund M, Madlensky L, et al. Breast cancer screening in the precision medicine era: risk-based screening in. population-based trial. J Natl Cancer Inst. 2017;109:djw290.
Acknowledgements
We thank the Cancéropole Nord-Ouest and the Institut National du Cancer (INCa) for funding. The FREX Consortium was supported by France Génomique National infrastructure, funded as part of the “Investissement d’Avenir” program managed by Agence Nationale pour la Recherche (contrat ANR-10-INBS-09). The French National League Against Cancer (LNC) and the French National Cancer Institute (INCa) provided grants to the Northwest Data Center (CTD-CNO). We also thank the high-throughput sequencing platform of Basse-Normandie SéSAME (Sequencing for Health, Agronomy, the Sea and the Environment) for technological support. We are grateful to Camille Charbonnier-Le Clezio for critical review of the manuscript.
Principal coinvestigators of the French Exome Project consortium
Emmanuelle Génin, Richard Redon, Jean-François Deleuze, Dominique Campion, Jean-Charles Lambert, and Jean-François Dartiques.
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
DISCLOSURE
The authors declare no conflict of interest.
Additional information
Member's of the French Exome Project Consortium are listed below Acknowledgement.
Rights and permissions
About this article
Cite this article
Castéra, L., Harter, V., Muller, E. et al. Landscape of pathogenic variations in a panel of 34 genes and cancer risk estimation from 5131 HBOC families. Genet Med 20, 1677–1686 (2018). https://doi.org/10.1038/s41436-018-0005-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-018-0005-9
Keywords
This article is cited by
-
Clinical characteristics and survival analysis of Chinese ovarian cancer patients with RAD51D germline mutations
BMC Cancer (2022)
-
Increased incidence of pathogenic variants in ATM in the context of testing for breast and ovarian cancer predisposition
Journal of Human Genetics (2022)
-
Germline sequence variants contributing to cancer susceptibility in South African breast cancer patients of African ancestry
Scientific Reports (2022)
-
First international workshop of the ATM and cancer risk group (4-5 December 2019)
Familial Cancer (2022)
-
Investigation of monogenic causes of familial breast cancer: data from the BEACCON case-control study
npj Breast Cancer (2021)