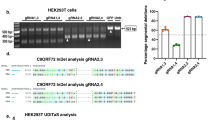
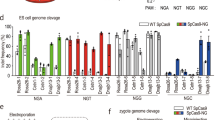
Abstract
Most Friedreich ataxia (FRDA) cases are caused by the elongation of the GAA repeat (GAAr) sequence in the first intron of the FXN gene, leading to a decrease of the frataxin protein expression. Deletion of this GAAr with CRISPR/Cas9 technology leads to an increase in frataxin expression in vitro. We are therefore aiming to develop FRDA treatment based on the deletion of GAAr with CRISPR/Cas9 technology using a single AAV expressing a small Cas9 (CjCas9) and two single guide RNAs (sgRNAs) targeting the FXN gene. This AAV was intraperitoneally administrated to YG8sR (250–300 GAAr) and to YG8-800 (800 GAAr) mice. DNA and RNA were extracted from different organs a month later. PCR amplification of part of intron 1 of the FXN gene detected some GAAr deletion in some cells in heart and liver of both mouse models, but the editing rate was not sufficient to cause an increase in frataxin mRNA in the heart. However, the correlation observed between the editing rate and the distribution of AAV suggests a possible therapy based on the removal of the GAAr with a better delivery tool of the CRISPR/Cas9 system.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data will be made available upon request.
References
Pandolfo M. Friedreich ataxia. In: PJ Vinken and GW Bruyn, editors. Handbook of clinical neurology. 2012;103:275–94.
Vankan P. Prevalence gradients of Friedreich’s ataxia and R1b haplotype in Europe co-localize, suggesting a common Palaeolithic origin in the Franco-Cantabrian ice age refuge. J Neurochem. 2013;126:11–20.
Pandolfo M. Friedreich ataxia: new pathways. J Child Neurol. 2012;27:1204–11.
Tsou AY, Paulsen EK, Lagedrost SJ, Perlman SL, Mathews KD, Wilmot GR, et al. Mortality in Friedreich ataxia. J Neurol Sci. 2011;307:46–9.
Pandolfo M. Friedreich ataxia: the clinical picture. J Neurol. 2009;256:3–8.
Galea CA, Huq A, Lockhart PJ, Tai G, Corben LA, Yiu EM, et al. Compound heterozygous FXN mutations and clinical outcome in friedreich ataxia. Ann Neurol. 2016;79:485–95.
Soragni E, Herman D, Dent SY, Gottesfeld JM, Wells RD, Napierala M. Long intronic GAA*TTC repeats induce epigenetic changes and reporter gene silencing in a molecular model of Friedreich ataxia. Nucleic Acids Res. 2008;36:6056–65.
Anzovino A, Lane DJ, Huang ML, Richardson DR. Fixing frataxin: ‘ironing out’ the metabolic defect in Friedreich’s ataxia. Br J Pharmacol. 2014;171:2174–90.
Lill R. Function and biogenesis of iron-sulphur proteins. Nature. 2009;460:831–8.
Perdomini M, Hick A, Puccio H, Pook MA. Animal and cellular models of Friedreich ataxia. J Neurochem. 2013;126:65–79.
Anjomani Virmouni S, Ezzatizadeh V, Sandi C, Sandi M, Al-Mahdawi S, Chutake Y, et al. A novel GAA-repeat-expansion-based mouse model of Friedreich’s ataxia. Dis Model Mech. 2015;8:225–35.
Ocana-Santero G, Diaz-Nido J, Herranz-Martin S. Future prospects of gene therapy for Friedreich’s ataxia. Int J Mol Sci. 2021;22:1815.
Gerard C, Archambault AF, Bouchard C, Tremblay JP. A promising mouse model for Friedreich ataxia progressing like human patients. Behav Brain Res. 2022;436:114107.
Ouellet DL, Cherif K, Rousseau J, Tremblay JP. Deletion of the GAA repeats from the human frataxin gene using the CRISPR-Cas9 system in YG8R-derived cells and mouse models of Friedreich ataxia. Gene Ther. 2017;24:265–74.
Kotterman MA, Schaffer DV. Engineering adeno-associated viruses for clinical gene therapy. Nat Rev Genet. 2014;15:445–51.
He X, Urip BA, Zhang Z, Ngan CC, Feng B. Evolving AAV-delivered therapeutics towards ultimate cures. J Mol Med (Berl). 2021;99:593–617.
Kim E, Koo T, Park SW, Kim D, Kim K, Cho HY, et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat commun. 2017;8:14500.
Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–32.
Ding X, Seebeck T, Feng Y, Jiang Y, Davis GD, Chen F. Improving CRISPR-Cas9 genome editing efficiency by fusion with chromatin-modulating peptides. CRISPR J. 2019;2:51–63.
Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168.
Mazzara PG, Muggeo S, Luoni M, Massimino L, Zaghi M, Valverde PT, et al. Frataxin gene editing rescues Friedreich’s ataxia pathology in dorsal root ganglia organoid-derived sensory neurons. Nat Commun. 2020;11:4178.
Li Y, Polak U, Bhalla AD, Rozwadowska N, Butler JS, Lynch DR, et al. Excision of Expanded GAA Repeats Alleviates the Molecular Phenotype of Friedreich’s Ataxia. Mol Ther. 2015;23:1055–65.
Li J, Rozwadowska N, Clark A, Fil D, Napierala JS, Napierala M. Excision of the expanded GAA repeats corrects cardiomyopathy phenotypes of iPSC-derived Friedreich’s ataxia cardiomyocytes. Stem Cell Res. 2019;40:101529.
Li Y, Li J, Wang J, Zhang S, Giles K, Prakash TP, et al. Premature transcription termination at the expanded GAA repeats and aberrant alternative polyadenylation contributes to the Frataxin transcriptional deficit in Friedreich’s ataxia. Hum Mol Genet. 2022;31:3539–57.
Sivakumar A, Cherqui S. Advantages and limitations of gene therapy and gene editing for Friedreich’s ataxia. Front Genome Ed. 2022;4:903139.
Rocca CJ, Rainaldi JN, Sharma J, Shi Y, Haquang JH, Luebeck J, et al. CRISPR-Cas9 gene editing of hematopoietic stem cells from patients with Friedreich’s ataxia. Mol Ther Methods Clin Dev. 2020;17:1026–36.
Belbellaa B, Reutenauer L, Monassier L, Puccio H. Correction of half the cardiomyocytes fully rescue Friedreich ataxia mitochondrial cardiomyopathy through cell-autonomous mechanisms. Hum Mol Genet. 2019;28:1274–85.
Belbellaa B, Reutenauer L, Messaddeq N, Monassier L, Puccio H. High levels of Frataxin overexpression lead to mitochondrial and cardiac toxicity in mouse models. Mol Ther Methods Clin Dev. 2020;19:120–38.
Li L, Matsui M, Corey DR. Activating frataxin expression by repeat-targeted nucleic acids. Nat commun. 2016;7:10606.
Greene E, Mahishi L, Entezam A, Kumari D, Usdin K. Repeat-induced epigenetic changes in intron 1 of the frataxin gene and its consequences in Friedreich ataxia. Nucleic Acids Res. 2007;35:3383–90.
Li K, Singh A, Crooks DR, Dai X, Cong Z, Pan L, et al. Expression of human frataxin is regulated by transcription factors SRF and TFAP2. PLoS One. 2010;5:e12286.
Yameogo P, Duchene BL, Majeau N, Tremblay JP. CRISPR-SCReT (CRISPR-Stop Codon Read Through) method to control Cas9 expression for gene editing. Gene Ther. 2022;29:171–7.
Yamada M, Watanabe Y, Gootenberg JS, Hirano H, Ran FA, Nakane T, et al. Crystal structure of the minimal Cas9 from campylobacter jejuni reveals the molecular diversity in the CRISPR-Cas9 Systems. Mol Cell. 2017;65:1109–21.
Silva-Pinheiro P, Cerutti R, Luna-Sanchez M, Zeviani M, Viscomi C. A single intravenous injection of AAV-PHP.B-hNDUFS4 ameliorates the phenotype of Ndufs4 (−/−) Mice. Mol Ther Methods Clin Dev. 2020;17:1071–8.
Chapdelaine P, Gerard C, Sanchez N, Cherif K, Rousseau J, Ouellet DL, et al. Development of an AAV9 coding for a 3XFLAG-TALEfrat#8-VP64 able to increase in vivo the human frataxin in YG8R mice. Gene Ther. 2016;23:606–14.
Tabebordbar M, Lagerborg KA, Stanton A, King EM, Ye S, Tellez L, et al. Directed evolution of a family of AAV capsid variants enabling potent muscle-directed gene delivery across species. Cell. 2021;184:4919–38.
Rocca CJ, Goodman SM, Dulin JN, Haquang JH, Gertsman I, Blondelle J, et al. Transplantation of wild-type mouse hematopoietic stem and progenitor cells ameliorates deficits in a mouse model of Friedreich’s ataxia. Sci Transl Med. 2017;9:eaaj2347.
Park H, Shin J, Choi H, Cho B, Kim J. Valproic acid significantly improves CRISPR/Cas9-mediated gene editing. Cells. 2020;9:1447.
Chan PK, Torres R, Yandim C, Law PP, Khadayate S, Mauri M, et al. Heterochromatinization induced by GAA-repeat hyperexpansion in Friedreich’s ataxia can be reduced upon HDAC inhibition by vitamin B3. Hum Mol Genet. 2013;22:2662–75.
Rodden LN, Gilliam KM, Lam C, Rojsajjakul T, Mesaros C, Dionisi C, et al. DNA methylation in Friedreich ataxia silences expression of frataxin isoform E. Sci Rep. 2022;12:5031.
Acknowledgements
We thank Dre Chantal Guillemette for ddPCR equipment and Julie Carbonneau for her technical assistance. PY has been supported by a fellowship from the Canadian Francophonie Scholarship Program (CFSP).
Funding
This research project was supported by grants from the Canadian Institute of Health Research, ThéCel network and Ataxia Canada.
Author information
Authors and Affiliations
Contributions
PY designed and performed the experiments and wrote the manuscript. CG provided technical assistance for the experiments and corrected the manuscript. NM assisted with the design of the experiments and corrected the manuscript. JPT conceived the experiments and corrected the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yaméogo, P., Gérard, C., Majeau, N. et al. Removal of the GAA repeat in the heart of a Friedreich’s ataxia mouse model using CjCas9. Gene Ther 30, 612–619 (2023). https://doi.org/10.1038/s41434-023-00387-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41434-023-00387-0