Abstract
Objectives
To delineate the disease-causing mutations of the Stargardt disease-related genes in Chinese patients diagnosed with Stargardt disease or retinitis pigmentosa (RP) by whole exome sequencing analysis.
Methods
A total of 123 sporadic RP or Stargardt disease patients and 2 Stargardt disease families were recruited. All sporadic patients and the probands of the families were subjected to whole exome sequencing analysis. The candidate mutations were verified by direct sequencing based on the cosegregation pattern and in 200 control subjects and by the bioinformatics analyses.
Results
A total of three reported ABCA4 mutations were identified in the probands of the two Stargardt disease families. The probands and the affected family members with either homozygous or compound heterozygous mutations showed typical Stargardt disease features, which was absent in their unaffected family members. The cosegregation pattern confirmed the mode of recessive inheritance. Moreover, two sporadic Stargardt disease patients were identified to carry two novel ABCA4 and one PROM1 mutations. In addition, 13 novel variants were found in 119 sporadic RP patients in 7 Stargardt disease-related genes, and 8 novel missense variants were conserved across different species and predicted to be damaging to the protein. All 15 novel variants were absent in our 200 control subjects.
Conclusions
This study revealed 22.4% study subjects carrying Stargardt disease-related gene mutations with total 15 novel variants in seven Stargardt disease-related genes, assuring that targeted next-generation sequencing analysis is a high throughput strategy to facilitate the clinical diagnosis from suspicious patients and recommended as a routine examination for inherited retinal dystrophies.
Similar content being viewed by others
Introduction
Stargardt’s macular dystrophy (Stargardt disease (STGD)) is the most common form of inherited juvenile macular degeneration, with an estimated annual incidence of 0.110–0.128 per 100,000 individuals in the UK [1]. Clinical hallmarks of STGD are the macular atrophy and the yellow-white lipofuscin flecks at retinal pigment epithelium (RPE) [2]. RPE atrophy accompanies with progressive degeneration of photoreceptor cells, leading to irreversible central vision loss [3]. Visual acuity loss in STGD patients usually develops in the first or second decades of their life, and individuals developing STGD at a younger age have poorer visual prognosis than those with a later disease onset [4]. Herein, early identification of STGD mutations can facilitate the genetic counseling and initiate potential therapeutic treatments in advance [5].
Multiple genes have been reported to be associated with STGD. STGD 1 (STGD-1, Online Mendelian Inheritance in Man (OMIM)#248200) is inherited in autosomal recessive manner with the mutations in ATP-binding cassette subfamily A member 4 (ABCA4) [6] or cyclic nucleotide gated channel beta 3 (CNGB3) [7] gene, whereas STGD-3 (OMIM#600110) and STGD-4 (OMIM#603786) are inherited in autosomal dominant manner with the mutations in elongation of very long chain fatty acid elongase 4 (ELOVL4) [8] and prominin 1 (PROM1) [9] genes, respectively. Besides, mutations in peripherin 2 (PRPH2) [10], crumbs 1 (CRB1) [11], phosphodiesterase 6H (PDE6H) [12], and bestrophin 1 (BEST1) [13] have also been reported to be related to STGD. In Chinese populations, STGD in 60% of patients are caused by ABCA4 mutations [14, 15]. PROM1 mutation was also reported in a Chinese family with autosomal dominant STGD [16]. Yet, ELOVL4, PRPH2, and BEST1 mutations were not found in Chinese patients with STGD [14, 17, 18]. Critically, the STGD-related gene variants not only attribute to the phenotypes of classical STGD but also contribute to cone-rod dystrophy and retinitis pigmentosa (RP)-like phenotypes [19], broadening the genetic complexity of STGD. In this study, we aimed to determine the STGD-related gene mutations in a cohort of Chinese patients diagnosed with STGD and RP.
Materials and methods
Study subjects
The study protocol was approved by the Ethics Committee on Human Medical Research at the Joint Shantou International Eye Center of Shantou University and The Chinese University of Hong Kong, which is in accordance with the tenets of the Declaration of Helsinki. Written informed consents were obtained from all study subjects or their legal representatives after explanation of the nature and possible consequences of the study. A total of 119 sporadic RP and 4 sporadic STGD patients as well as 2 STGD families were recruited at the Joint Shantou International Eye Center of Shantou University and The Chinese University of Hong Kong. Comprehensive ophthalmic examinations, including best-corrected visual acuity, fundus photography, optical coherence tomography (OCT), and electroretinogram (ERG), were performed for all patients and some of the unaffected family members. The sporadic RP patients generally showed pubertal night blindness, restricted peripheral vision, progressive vision loss, overall bone-spicule pigmentation of the retina, attenuation of retinal vessels, and the flattening of the rod and cone ERG responses.
For the validation analysis, 200 unrelated control subjects, aged 60 years or above, were also enrolled. The control subjects did not have any family history or symptom of inherited retinal diseases or any other major eye diseases except mild senile cataracts or mild myopia.
Peripheral blood was collected from all study subjects, and genomic DNA was extracted by TIANGEN DNA blood Kit DP318 (TIANGEN, Beijing, China), and stored at −80 °C freezer before sequencing analysis.
Whole exome sequencing analysis
Whole exome sequencing analysis was conducted for the probands of the two recruited families and 123 sporadic patients according to our previous established protocols [20]. Briefly, genomic DNA was fragmented into 150–200 bp DNA libraries by S2/E210 Focused-ultrasonicator (Covaris, Woburn, MA). All coding exons were captured by and hybridized to a customized array (Agilent Technologies, Santa Clara, CA). The purified and enriched DNA libraries were sequenced using the HiSeq2500 platform (Illumina, San Diego, CA) to generate paired-end reads for 90 cycles per reads. The results were aligned to human genome reference (UCSC hg19) in the National Center for Biotechnology Information database using Burrows Wheeler Aligner Multi-Vision software package (BWA) for unique mapped reads. The single-nucleotide polymorphisms (SNPs) and insertion/deletion variants (indels) were identified using the SAMtools and BCFtools. The detected variants were annotated with ANNOVAR with reference to the following databases for further variant filtering, including 1000 Genome Project (http://www.internationalgenome.org/), gnomAD (http://gnomad-old.broadinstitute.org/), EXAC (http://exac.broadinstitute.org/), and dbSNP (http://www3.ncbi.nlm.nih.gov/SNP/).
To ensure the accuracy and efficacy of the candidate mutations, the synonymous, intergenic, and intronic variants were first filtered out. Those variants with minor allele frequency (MAF) lower than 1% or absent in East Asian population from gnomAD and EXAC were reserved. The candidate variants were compared to the 13 recommended testing panel genes for STGD (ABCA4, BEST1, C1QTNF5, CDH3, CNGB3, CRB1, ELOVL4, PROM1, PRPH2, RIMS1, RP1L1, RPGR, and TIMP3; https://bredagenetics.com/stargardt-disease/) [6,7,8,9,10,11,12,13]. The potential influence of the variants on protein structure/function was predicted by SIFT (http://sift.jcvi.org/), PolyPhen2 (http://genetics.bwh.harvard.edu/pph2/), MutationTaster (http://www.mutationtaster.org/), and CADD (http://cadd.gs.washington.edu/). ExPASy (https://web.expasy.org/translate/) was applied to predict the influence of the indel variants on the amino acid translation. Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) was used to conduct multiple sequence alignment to evaluate the conservation of the amino acid residues of the variants across different species.
Sanger sequencing validation
The putative gene variants identified by whole exome sequencing analysis were verified in family members and 200 control subjects by Sanger sequencing with specific primers (AIJI Biotechnology Company, Guangdong, China) using the ABI 3730XL Genetic Analyzer (Applied Biosystems, Foster City, CA) to confirm the cosegregation pattern. The sequencing results were analyzed by NOVOSNP (http://www.molgen.vib-ua.be/bioinfo/novosnp/) with the alignment to the reference DNA sequence.
Results
Whole exome sequencing analysis identified ABCA4 mutations in STGD families
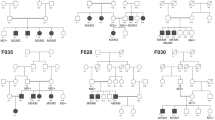
The two recruited Chinese families were diagnosed with STGD, and all followed the mode of recessive inheritance. In STGD family 1 (Fig. 1A), the parents (I:1 and I:2) were unaffected and did not show any clinical manifestations of STGD. In contrast, the proband (II:4) and her elder sister (II:1) first visited the outpatient clinic at the age of 8 and 9 years, respectively. Fundus examination identified yellowish flecks with reduced reflection in the macula, and OCT examination showed decreased macular thickness (Fig. 1C). Besides, the ERG responses were also flattened, collectively indicating the diagnosis of STGD. The remaining unaffected subjects (II:2, II:3, II:5, and II:6) did not show any symptoms or abnormalities in the ophthalmic examinations.
The pedigrees of the two recruited families diagnosed with Stargardt disease following the pattern of autosomal recessive inheritance: A family 1 and B family 2. Squares and circles represent men and women, respectively. Black: the affected members; white: the unaffected members. C The clinical characteristics of the probands of the two recruited Stargardt disease families, including color fundus photographs showing yellowish flecks and reduced reflection, reduced macular thickness in the OCT examination, and the flattened or delayed ERG signals as compared to the normal subject.
STGD family 2 is a consanguineous marriage (Fig. 1B). The parents (I:1 and I:2) did not show any symptoms or abnormalities in the ophthalmic examinations. However, the two affected children (the proband II:1, and II:2) first visited the outpatient clinic at the age of 15 and 11 years, respectively. Yellowish flecks and reduced reflection were observed in the macula in the fundus examination (Fig. 1C). The macular thickness was found to be reduced in the OCT examination. Besides, the ERG signals were also delayed. Collectively, the diagnosis of STGD was revealed.
The probands of the two recruited STGD families (family 1: II:4; family 2: II:1) were subjected to whole exome sequencing analysis. Each whole exome sequencing analysis resulted in a total of 70 GB of sequence data, and 95.5% of sequence reads were originated from exons, with a mean coverage of 100-fold. The total numbers of variants (SNPs and indels) of exons and splice sites identified were 23,501 for the proband of STGD family 1 and 23,338 for the proband of family 2 (Supplementary Fig. 1). After filtering the synonymous, intergenic, intronic, and common variants, the candidate deleterious variants in STGD-related genes with MAF < 1% and recessive inheritance were reduced to 2 for STGD family 1 and 1 for family 2.
For STGD family 1, two heterozygous variants in ABCA4 gene, a missense variant c.1804C>T and a splice site variant c.5196+1G>A (Table 1), were identified only in the two affected subjects (II:1 and the proband II:4). The identified variants have been confirmed by Sanger sequencing (Fig. 2). Their unaffected father (I:1) and sister (II:3) only carried the c.1804C>T variant, whereas the unaffected mother (I:2), a sister (II:2) and a brother (II:5) only carried the c.5196+1G>A variant. The younger brother (II:6) did not carry any variants. The transmission of the two identified variants followed the cosegregation pattern with STGD in this family (Fig. 1A). Both variants have been previously reported [14, 21], but not found in 200 control subjects from our cohort. Therefore, the c.1804C>T and c.5196+1G>A variants should be the causative mutations for this STGD family.
For STGD family 2, homozygous variant in ABCA4 gene, missense variant c.2894A>G in exon 19 (Table 1), was identified in the two affected subjects (the proband II:1, and II:2). The identified variants have been confirmed by Sanger sequencing (Fig. 2). Their unaffected father (I:1) and mother (I:2) each carried only one heterozygous c.2894A>G variant, following the cosegregation pattern with the disease phenotype transmission in this family (Fig. 1B). This variant has been previously reported [22], and was not found in 200 control subjects from our cohort. Therefore, homozygous c.2894A>G variant should be the causative mutations for this family with consanguineous marriage.
Whole exome sequencing analysis identified STGD-related gene variants in sporadic STGD and RP patients
To further extend the spectrum of STGD-related gene mutations, whole exome sequencing analysis was conducted in 4 sporadic STGD patients and 119 sporadic RP patients. Among the STGD patients, two novel heterozygous ABCA4 variants were identified in STGD-4 (c.4555delA and c.6397T>C; Table 1). Moreover, one reported (c.1196G>A) and one novel heterozygous PROM1 variants (c.1520T>A) were identified in STGD-1. Furthermore, one reported heterozygous ABCA4 missense variant (c.4610C>T) and two reported heterozygous RP1L1 missense variants (c.670G>A and c.6431A>C) were identified in STGD-3. In STGD-2, one reported heterozygous nonsense variant (c.5641G>T) and two reported heterozygous missense variants (c.5626G>A and c.5648C>T) were identified in RP1L1 gene. The three novel variants in ABCA4 and PROM1 genes were verified by Sanger sequencing (Fig. 2) and were not found in our 200 control subjects. Collectively, the ABCA4 and PROM1 variants could be the disease-causing mutations for STGD-4 and STGD-1 respectively, whereas RP1L1 variants could be associated with STGD for STGD-2 and STGD-3.
Among the 119 sporadic RP patients, total 69 variants were identified, and 13 of them were novel (Table 2). For the novel variants, four were identified in ABCA4 gene (c.53G>C, c.2054delC, c.6397T>C, and c.6479+1G>C), one in BEST1 gene (c.362G>C), one in CDH3 gene (c.1120G>A), two in CRB1 gene (c.2210T>C and c.3862G>A), two in RIMS1 gene (c.2T>C and c.228G>T), one in RP1L1 gene (c.413dupC), and two in RPGR gene (c.946A>T and c.3147delG). All these newly identified variants have been verified by Sanger sequencing (Fig. 2) and were not found in our 200 control subjects.
Bioinformatics analysis of STGD-related gene mutations
Among the 73 variants identified in this study (Tables 1 and 2), 15 variants were novel, including 10 missense variants, 4 indel variants, and 1 splice site variant. Among these missense variants, the amino acid residues of the seven variants in five Stargardt-related genes (ABCA4 p.R18P and p.C2133R, BEST1 p.G121A, CDH3 p.G374R, CRB1 p.1288S, and RIMS1 p.M1T and p.Q76H) were completely or remarkably conserved across eight different species, whereas RPGR p.M316L was considerably conserved across six different species (Fig. 3), indicating that these missense variants could be critical for the protein function or structure of the Stargardt-related genes. In contrast, CRB1 p.I737T and PROM1 p. F507Y were not conserved across different species.
Protein sequences of the Stargardt-related genes in Homo sapiens, Pan troglodytes, Bos taurus, Sus scrofa, Rattus norvegicus, Mus musculus, Gallus gallus, Numida meleagris, Xenopus tropicalis, Danio rerio, Chanos chanos, Zootoca vivipara, Drosophila melanogaster, Podarcis muralis, Geotrypetes seraphini, Coturnix japonica, and Trachemys scripta elegans were aligned by Clustal Omega based on the availability of the protein sequences. Multiple sequence alignment of the ten novel missense variants of the Stargardt-related genes was displayed.
The novel splice site variant in ABCA4 (c.6479+1G>C) would affect mRNA processing of ABCA4 gene at exon 47, which is expected to form a truncated protein or induce the nonsense-mediated decay. Furthermore, the novel indel variants were predicted to induce drastic changes to the protein structure with translational frameshifting and premature stop codon (ABCA4 c.2054delC: premature protein with 685 in-frame amino acids only; ABCA4 c.4555delA: premature protein with 1519 in-frame and 5 out-of-frame amino acids; RP1L1 c.413dupC: premature protein with 138 in-frame and 10 out-of-frame amino acids; RPGR c.3147delG: premature protein with 1049 in-frame and 38 out-of-frame amino acids).
Discussion
Inherited retinal dystrophies (IRDs) belong to a group of rare heterogeneous chronic disorders with degeneration of photoreceptors and RPE. As different subtypes of IRDs share similar phenotypes and these similar clinical manifestations could be caused by different gene mutations, ophthalmic examination alone might not easily specify the exact type of disorder. Currently, more than 200 genes have been discovered as the disease-causing genes for IRDs [21]. In this scenario, whole exome sequencing analysis could facilitate the precise genetic testing of these heterogeneous disorders for better clinical diagnosis, management, and counseling [20].
STGD could be caused by the mutations in multiple genes with different inheritance mode. ABCA4 and CNGB3 gene mutations are inherited in autosomal recessive manner [6, 7], whereas ELOVL4 and PROM1 gene mutations are inherited in autosomal dominant manner [8, 9]. For the Stargardt-related genes, mutations in PRPH2, PDE6H, and RIMS1 mutations are inherited in autosomal dominant manner [10, 12, 23], whereas CRB1, CDH3, and RP1L1 mutations are inherited in autosomal recessive manner [11, 24, 25] and RPGR is X-linked inheritance [26]. Besides, BEST1 mutations might increase the variation burden on the disease severity [13]. In this study, we identified 73 rare variants in 13 Stargardt-related genes among 2 STGD families as well as 4 sporadic STGD patients and 119 sporadic RP patients. Following the inheritance modes of the STGD-related genes, six study subjects carry homozygous or compound heterozygous ABCA4 variants, nine subjects with compound heterozygous RP1L1 variants, one sporadic RP patient with compound heterozygous CRB1 variants, three subjects with PROM1 variants, three sporadic RP patients with PRPH2 variants, three sporadic RP patients with RIMS1 variants, and three male sporadic RP patients with RPGR variants (Tables 1 and 2). Altogether, 28 out of 125 (22.4%) unrelated study subjects carry the potential disease-causing gene variants in six Stargardt-related genes. With the in-depth analysis, the percentage of sporadic RP patients (18.5%; 22 out of 119) carrying potential disease-causing Stargardt-related gene variants is higher than those carrying gene variants for Usher syndrome (9.2%; 11 out of 119) in our southern Chinese population [20].
Among the STGD families and sporadic patients, we identified homozygous or compound heterozygous ABCA4 variants in two STGD families and one sporadic STGD patient (Table 1), indicating that 50% of study subjects with STGD carry the ABCA4 mutations. This is comparable to the previous reports on STGD (60%) in Chinese populations [14, 15]. Moreover, we also identified two PROM1 variants (p.R399H and p.F507Y) in one sporadic STGD patient. PROM1 mutation has been reported in a Chinese family with autosomal dominant STGD as well as in sporadic RP patients [16, 27]. However, similar to other studies [14, 17, 18], ELOVL4, PRPH2, and BEST1 mutation was not found in our patients with STGD. Notably, two sporadic STGD patients carry compound heterozygous variants in the RP1L1 gene. Whether these two patients were affected by occult macular dystrophy requires further clinical examinations [28]. Nevertheless, this implicated the complexity of clinical manifestations that targeted next-generation sequencing could aid the molecular diagnosis of IRDs.
In this study, we identified 15 novel variants in 8 Stargardt-related genes (ABCA4, BEST1, CDH3, CRB1, PROM1, RIMS1, RP1L1, and RPGR), including 10 missense variants, 4 indel variants, and 1 splice site variant (Tables 1 and 2). All 15 novel variants were absent in our 200 control subjects. Among the novel missense variants, seven of them (ABCA4 p.R18P and p.C2133R, BEST1 p.G121A, CDH3 p.G374R, CRB1 p.1288S, and RIMS1 p.M1T and p.Q76H) are completely or remarkably conserved across eight different species, whereas RPGR p.M316L is considerably conserved across six different species (Fig. 3), indicating that these missense variants could be critical for the protein function or structure of the Stargardt-related genes. In contrast, CRB1 p.I737T is only conserved among Homo sapiens, Pan troglodytes, and Xenopus tropicalis, whereas PROM1 p. F507Y is conserved in Homo sapiens and Pan troglodytes, suggesting that these two variants might only be critical in primates. For the novel splice site variant, ABCA4 (c.6479+1G>C) variant could affect mRNA processing of ABCA4 gene, which is expected to form a truncated protein or induce the nonsense-mediated decay. Furthermore, the novel indel variants (ABCA4 c.2054delC and c.4555delA, RP1L1 c.413dupC and RPGR c.3147delG) are predicted to induce drastic changes to the protein structure with translational frameshifting and premature stop codon. Therefore, these Stargardt-related gene variants likely play a disease-causing role in STGD and RP. However, how these variants influence the protein function of the Stargardt-related genes requires further investigations.
ABCA4 belongs to the retina-specific ATP-binding cassette transporter and is expressed in the outer segment disc membranes of photoreceptor cells, responsible for the transportation of N-retinylidene-phosphatidylethanolamine from lumen leaflet to cytoplasmic surface of disc membranes so as to prevent the accumulation of retinal and its toxic bisretinoid compounds in RPE after phagocytosis of the detached outer segment [29]. ABCA4 protein is composed of two transmembrane domains, two glycosylated extracellular domains, and two nucleotide binding domains with ATPase activity [30]. ABCA4 gene mutations have been shown to attenuate the clearance rate of retinal in the disc membranes and the deposition of bisretinoid-lipofuscin in the visual cycle, which results in RPE degeneration [31]. PROM1 gene encodes a five-transmembrane domain glycoprotein with two large N-glycosylated extracellular loops involved in the formation and organization of outer segment disc membrane [32]. In Prom1-knockout mice, photoreceptor cell degeneration is light-dependent, and the retina shows downregulated expression of Rdh12 and Abca4 [33]. In zebrafish with loss of prom1b, outer segment morphogenesis is disrupted with cone degeneration at early age and rods remained viable but with abnormal outer segment [34]. Prom1b deletion also causes mislocalization of Prph2 and disrupts its oligomerization. RP1L1 gene encodes for a retinal-specific protein component of the photoreceptor cilium, which is essential for outer segment morphogenesis of photoreceptors [35]. Mutations in RP1L1 gene would result in remarkable photoreceptor disruption [36]. PRPH2, RPGR, and CRB1 genes encode proteins for the outer or inner segment of photoreceptors, and their mutations have been shown causing macular dystrophy and RP [37,38,39,40,41]. How the novel Stargardt-related gene variants could be involved in the disease-causing mechanisms warrants in-depth analyses in future studies.
Conclusions
In summary, this study revealed a total of 15 novel variants in seven STGD-related genes among the Chinese STGD and RP patients, expanding the mutation spectrum for STGD and RP. We recommended targeted next-generation sequencing analysis as a routine high throughput genetic examination to facilitate the clinical diagnosis of IRDs for the suspicious patients.
Summary
What was known before
-
STGD is associated at least with eight different genes.
-
ABCA4 mutations are the most common disease-causing mutations for STGD.
-
Whole exome sequencing analysis is a high throughput strategy to identify disease-causing mutations for IRDs.
What this study adds
-
This study identified 15 novel variants in eight Stargardt-related genes in Chinese patients diagnosed with STGD or RP.
-
18.5% sporadic RP patients carry STGD-related gene mutations.
-
Compound heterozygous mutations in RP1L1 gene are associated with STGD.
References
Spiteri Cornish K, Ho J, Downes S, Scott NW, Bainbridge J, Lois N. The epidemiology of Stargardt disease in the United Kingdom. Ophthalmol Retin. 2017;1:508–13.
Fujinami K, Zernant J, Chana RK, Wright GA, Tsunoda K, Ozawa Y, et al. Clinical and molecular characteristics of childhood-onset Stargardt disease. Ophthalmology. 2015;122:326–34.
Rotenstreich Y, Fishman GA, Anderson RJ. Visual acuity loss and clinical observations in a large series of patients with Stargardt disease. Ophthalmology. 2003;110:1151–8.
Westeneng-van Haaften SC, Boon CJ, Cremers FP, Hoefsloot LH, den Hollander AI, Hoyng CB. Clinical and genetic characteristics of late-onset Stargardt’s disease. Ophthalmology. 2012;119:1199–210.
Lu LJ, Liu J, Adelman RA. Novel therapeutics for Stargardt disease. Graefes Arch Clin Exp Ophthalmol. 2017;255:1057–62.
Allikmets R, Singh N, Sun H, Shroyer NF, Hutchinson A, Chidambaram A, et al. A photoreceptor cell-specific ATP-binding transporter gene (ABCR) is mutated in recessive Stargardt macular dystrophy. Nat Genet. 1997;15:236–46.
Corton M, Nishiguchi KM, Avila-Fernandez A, Nikopoulos K, Riveiro-Alvarez R, Tatu SD, et al. Exome sequencing of index patients with retinal dystrophies as a tool for molecular diagnosis. PLoS ONE. 2013;8:e65574.
Zhang K, Kniazeva M, Han M, Li W, Yu Z, Yang Z, et al. A 5-bp deletion in ELOVL4 is associated with two related forms of autosomal dominant macular dystrophy. Nat Genet. 2001;27:89–93.
Maw MA, Corbeil D, Koch J, Hellwig A, Wilson-Wheeler JC, Bridges RJ, et al. A frameshift mutation in prominin (mouse)-like 1 causes human retinal degeneration. Hum Mol Genet. 2000;9:27–34.
Boon CJ, van Schooneveld MJ, den Hollander AI, van Lith-Verhoeven JJ, Zonneveld-Vrieling MN, Theelen T, et al. Mutations in the peripherin/RDS gene are an important cause of multifocal pattern dystrophy simulating STGD1/fundus flavimaculatus. Br J Ophthalmol. 2007;91:1504–11.
Riveiro-Alvarez R, Vallespin E, Wilke R, Garcia-Sandoval B, Cantalapiedra D, Aguirre-Lamban J, et al. Molecular analysis of ABCA4 and CRB1 genes in a Spanish family segregating both Stargardt disease and autosomal recessive retinitis pigmentosa. Mol Vis. 2008;14:262–7.
Piri N, Gao YQ, Danciger M, Mendoza E, Fishman GA, Farber DB. A substitution of G to C in the cone cGMP-phosphodiesterase gamma subunit gene found in a distinctive form of cone dystrophy. Ophthalmology. 2005;112:159–66.
Zolnikova IV, Strelnikov VV, Skvortsova NA, Tanas AS, Barh D, Rogatina EV, et al. Stargardt disease-associated mutation spectrum of a Russian Federation cohort. Eur J Med Genet. 2017;60:140–7.
Xin W, Xiao X, Li S, Jia X, Guo X, Zhang Q. Identification of genetic defects in 33 probands with Stargardt disease by WES-based bioinformatics gene panel analysis. PLoS ONE. 2015;10:e0132635.
Jiang F, Pan Z, Xu K, Tian L, Xie Y, Zhang X, et al. Screening of ABCA4 gene in a Chinese cohort with Stargardt disease or cone-rod dystrophy with a report on 85 novel mutations. Investig Ophthalmol Vis Sci. 2016;57:145–52.
Imani S, Cheng J, Shasaltaneh MD, Wei C, Yang L, Fu S, et al. Genetic identification and molecular modeling characterization reveal a novel PROM1 mutation in Stargardt4-like macular dystrophy. Oncotarget. 2018;9:122–41.
Lai Z, Zhang XN, Zhou W, Yu R, Le YP. Evaluation of the ELOVL4 gene in a Chinese family with autosomal dominant STGD3-like macular dystrophy. J Cell Mol Med. 2005;9:961–5.
Yi J, Li S, Jia X, Xiao X, Wang P, Guo X, et al. Evaluation of the ELOVL4, PRPH2 and ABCA4 genes in patients with Stargardt macular degeneration. Mol Med Rep. 2012;6:1045–9.
Cremers FPM, Lee W, Collin RWJ, Allikmets R. Clinical spectrum, genetic complexity and therapeutic approaches for retinal disease caused by ABCA4 mutations. Prog Retin Eye Res. 2020;79:100861.
Ng TK, Tang W, Cao Y, Chen S, Zheng Y, Xiao X, et al. Whole exome sequencing identifies novel USH2A mutations and confirms Usher syndrome 2 diagnosis in Chinese retinitis pigmentosa patients. Sci Rep. 2019;9:5628.
Ran X, Cai WJ, Huang XF, Liu Q, Lu F, Qu J, et al. ‘RetinoGenetics’: a comprehensive mutation database for genes related to inherited retinal degeneration. Database. 2014;2014:bau047.
Rosenberg T, Klie F, Garred P, Schwartz M. N965S is a common ABCA4 variant in Stargardt-related retinopathies in the Danish population. Mol Vis. 2007;13:1962–9.
Sisodiya SM, Thompson PJ, Need A, Harris SE, Weale ME, Wilkie SE, et al. Genetic enhancement of cognition in a kindred with cone-rod dystrophy due to RIMS1 mutation. J Med Genet. 2007;44:373–80.
Albarry MA, Hashmi JA, Alreheli AQ, Albalawi AM, Khan B, Ramzan K, et al. Novel homozygous loss-of-function mutations in RP1 and RP1L1 genes in retinitis pigmentosa patients. Ophthalmic Genet. 2019;40:507–13.
Khan AO, Bolz HJ. Phenotypic observations in “hypotrichosis with juvenile macular dystrophy” (recessive CDH3 mutations). Ophthalmic Genet. 2016;37:301–6.
Cehajic-Kapetanovic J, McClements ME, Whitfield J, Shanks M, Clouston P, MacLaren RE. Association of a novel intronic variant in RPGR with hypomorphic phenotype of X-linked retinitis pigmentosa. JAMA Ophthalmol. 2020;138:1151–8.
Liu S, Xie L, Yue J, Ma T, Peng C, Qiu B, et al. Whole-exome sequencing identifies a novel homozygous frameshift mutation in the PROM1 gene as a causative mutation in two patients with sporadic retinitis pigmentosa. Int J Mol Med. 2016;37:1528–34.
Qi YH, Gao FJ, Hu FY, Zhang SH, Chen JY, Huang WJ, et al. Next-generation sequencing-aided rapid molecular diagnosis of occult macular dystrophy in a Chinese family. Front Genet. 2017;8:107.
Molday RS. Insights into the molecular properties of ABCA4 and its role in the visual cycle and Stargardt disease. Prog Mol Biol Transl Sci. 2015;134:415–31.
Quazi F, Molday RS. Differential phospholipid substrates and directional transport by ATP-binding cassette proteins ABCA1, ABCA7, and ABCA4 and disease-causing mutants. J Biol Chem. 2013;288:34414–26.
Zhao J, Liao Y, Chen J, Dong X, Gao Z, Zhang H, et al. Aberrant buildup of all-trans-retinal dimer, a nonpyridinium bisretinoid lipofuscin fluorophore, contributes to the degeneration of the retinal pigment epithelium. Investig Ophthalmol Vis Sci. 2017;58:1063–75.
Weigmann A, Corbeil D, Hellwig A, Huttner WB. Prominin, a novel microvilli-specific polytopic membrane protein of the apical surface of epithelial cells, is targeted to plasmalemmal protrusions of non-epithelial cells. Proc Natl Acad Sci USA. 1997;94:12425–30.
Dellett M, Sasai N, Nishide K, Becker S, Papadaki V, Limb GA, et al. Genetic background and light-dependent progression of photoreceptor cell degeneration in prominin-1 knockout mice. Investig Ophthalmol Vis Sci. 2014;56:164–76.
Lu Z, Hu X, Reilly J, Jia D, Liu F, Yu S, et al. Deletion of the transmembrane protein Prom1b in zebrafish disrupts outer-segment morphogenesis and causes photoreceptor degeneration. J Biol Chem. 2019;294:13953–63.
Noel NCL, MacDonald IM. RP1L1 and inherited photoreceptor disease: a review. Surv Ophthalmol. 2020;65:725–39.
Ahn SJ, Cho SI, Ahn J, Park SS, Park KH, Woo SJ. Clinical and genetic characteristics of Korean occult macular dystrophy patients. Investig Ophthalmol Vis Sci. 2013;54:4856–63.
Strafella C, Caputo V, Pagliaroli G, Iozzo N, Campoli G, Carboni S, et al. NGS analysis for molecular diagnosis of retinitis pigmentosa (RP): detection of a novel variant in PRPH2 gene. Genes. 2019;10:792.
Khan KN, Robson A, Mahroo OAR, Arno G, Inglehearn CF, Armengol M, et al. A clinical and molecular characterisation of CRB1-associated maculopathy. Eur J Hum Genet. 2018;26:687–94.
Guo X, Li J, Wang Q, Shu Y, Wang J, Chen L, et al. Identification of CRB1 mutations in two Chinese consanguineous families exhibiting autosomal recessive retinitis pigmentosa. Mol Med Rep. 2019;20:2922–8.
Koyanagi Y, Ueno S, Ito Y, Kominami T, Komori S, Akiyama M, et al. Relationship between macular curvature and common causative genes of retinitis pigmentosa in Japanese patients. Investig Ophthalmol Vis Sci. 2020;61:6.
Birtel J, Eisenberger T, Gliem M, Muller PL, Herrmann P, Betz C, et al. Clinical and genetic characteristics of 251 consecutive patients with macular and cone/cone-rod dystrophy. Sci Rep. 2018;8:4824.
Acknowledgements
The authors would like to express their deepest gratitude to all the participants in this study.
Funding
This work was supported by the Special Fund for the Innovative Science and Technology Strategy of Guangdong Province (project code: 180918154960752 to TKN), an internal grant from the Joint Shantou International Eye Center of Shantou University and The Chinese University of Hong Kong (project code: 20-020 to TKN), the National Nature Science Foundation of China (30901646 and 81170853 to HC), YangFan Program and TeZhi Program of Guangdong Province (to HC), and Grant for Key Disciplinary Project of Clinical Medicine under the Guangdong High-level University Development Program, China.
Author information
Authors and Affiliations
Contributions
TKN and HC: conception and design and financial support. YZ and HC: provision of study materials. YC, X-LY, SC, YX, and S-LC: collection and/or assembly of data. TKN, YC, and X-LY: data analysis and interpretation. TKN and X-LY: manuscript writing. TKN and HC: final approval of manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Ng, T.K., Cao, Y., Yuan, XL. et al. Whole exome sequencing analysis identifies novel Stargardt disease-related gene mutations in Chinese Stargardt disease and retinitis pigmentosa patients. Eye 36, 749–759 (2022). https://doi.org/10.1038/s41433-021-01525-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41433-021-01525-x