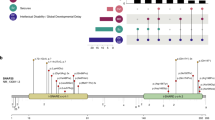
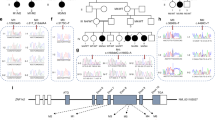
Abstract
Translation elongation factor eEF1A2 constitutes the alpha subunit of the elongation factor-1 complex, responsible for the enzymatic binding of aminoacyl-tRNA to the ribosome. Since 2012, 21 pathogenic missense variants affecting EEF1A2 have been described in 42 individuals with a severe neurodevelopmental phenotype including epileptic encephalopathy and moderate to profound intellectual disability (ID), with neurological regression in some patients. Through international collaborative call, we collected 26 patients with EEF1A2 variants and compared them to the literature. Our cohort shows a significantly milder phenotype. 83% of the patients are walking (vs. 29% in the literature), and 84% of the patients have language skills (vs. 15%). Three of our patients do not have ID. Epilepsy is present in 63% (vs. 93%). Neurological examination shows a less severe phenotype with significantly less hypotonia (58% vs. 96%), and pyramidal signs (24% vs. 68%). Cognitive regression was noted in 4% (vs. 56% in the literature). Among individuals over 10 years, 56% disclosed neurocognitive regression, with a mean age of onset at 2 years. We describe 8 novel missense variants of EEF1A2. Modeling of the different amino-acid sites shows that the variants associated with a severe phenotype, and the majority of those associated with a moderate phenotype, cluster within the switch II region of the protein and thus may affect GTP exchange. In contrast, variants associated with milder phenotypes may impact secondary functions such as actin binding. We report the largest cohort of individuals with EEF1A2 variants thus far, allowing us to expand the phenotype spectrum and reveal genotype-phenotype correlations.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
All anonymized data and related documentation from this study are available on reasonable request. Declaration to public database: All variants are reported and annotated in ClinVar website (accession number SCV004171535- SCV004171551): https://www.ncbi.nlm.nih.gov/clinvar/.
Change history
03 April 2024
A Correction to this paper has been published: https://doi.org/10.1038/s41431-024-01606-x
References
Kahns S, Lund A, Kristensen P, Knudsen CR, Clark BF, Cavallius J, et al. The elongation factor 1 A-2 isoform from rabbit: cloning of the cDNA and characterization of the protein. Nucleic Acids Res. 1998;26:1884–90. https://doi.org/10.1093/nar/26.8.1884.
Abbott CM, Newbery HJ, Squires CE, Brownstein D, Griffiths LA, Soares DC. eEF1A2 and neuronal degeneration. Biochem Soc Trans. 2009;37:1293–7. https://doi.org/10.1042/BST0371293.
Newbery HJ, Loh DH, O’Donoghue JE, Tomlinson VAL, Chau Y-Y, Boyd JA, et al. Translation elongation factor eEF1A2 is essential for post-weaning survival in mice. J Biol Chem. 2007;282:28951–9. https://doi.org/10.1074/jbc.M703962200.
Knudsen SM, Frydenberg J, Clark BF, Leffers H. Tissue-dependent variation in the expression of elongation factor-1 alpha isoforms: isolation and characterisation of a cDNA encoding a novel variant of human elongation-factor 1 alpha. Eur J Biochem. 1993;215:549–54. https://doi.org/10.1111/j.1432-1033.1993.tb18064.x.
Chambers DM, Peters J, Abbott CM. The lethal mutation of the mouse wasted (wst) is a deletion that abolishes expression of a tissue-specific isoform of translation elongation factor 1alpha, encoded by the Eef1a2 gene. Proc Natl Acad Sci USA. 1998;95:4463–8. https://doi.org/10.1073/pnas.95.8.4463.
Khalyfa A, Carlson BM, Dedkov EI, Wang E. Changes in protein levels of elongation factors, eEF1A-1 and eEF1A-2/S1, in long-term denervated rat muscle. Restor Neurol Neurosci. 2003;21:47–53.
McLachlan F, Sires AM, Abbott CM. The role of translation elongation factor eEF1 subunits in neurodevelopmental disorders. Hum Mutat. 2019;40:131–41. https://doi.org/10.1002/humu.23677.
De Ligt J, Willemsen MH, van Bon BWM, Kleefstra T, Yntema HG, Kroes T, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N. Engl J Med. 2012;367:1921–9. https://doi.org/10.1056/NEJMoa1206524.
Carvill GL, Helbig KL, Myers CT, Scala M, Huether R, Lewis S, et al. Damaging de novo missense variants in EEF1A2 lead to a developmental and degenerative epileptic-dyskinetic encephalopathy. Hum Mutat. 2020;41:1263–79. https://doi.org/10.1002/humu.24015.
Veeramah KR, Johnstone L, Karafet TM, Wolf D, Sprissler R, Salogiannis J, et al. Exome sequencing reveals new causal mutations in children with epileptic encephalopathies. Epilepsia. 2013;54:1270–81. https://doi.org/10.1111/epi.12201.
Long K, Wang H, Song Z, Yin X, Wang Y. EEF1A2 mutations in epileptic encephalopathy/intellectual disability: Understanding the potential mechanism of phenotypic variation. Epilepsy Behav EB. 2020;105:106955. https://doi.org/10.1016/j.yebeh.2020.106955.
Inui T, Kobayashi S, Ashikari Y, Sato R, Endo W, Uematsu M, et al. Two cases of early-onset myoclonic seizures with continuous parietal delta activity caused by EEF1A2 mutations. Brain Dev. 2016;38:520–4. https://doi.org/10.1016/j.braindev.2015.11.003.
Cao S, Smith LL, Padilla-Lopez SR, Guida BS, Blume E, Shi J, et al. Homozygous EEF1A2 mutation causes dilated cardiomyopathy, failure to thrive, global developmental delay, epilepsy and early death. Hum Mol Genet. 2017;26:3545–52. https://doi.org/10.1093/hmg/ddx239.
Kaneko M, Rosser T, Raca G. Dilated cardiomyopathy in a patient with autosomal dominant EEF1A2-related neurodevelopmental disorder. Eur J Med Genet. 2021;64:104121. https://doi.org/10.1016/j.ejmg.2020.104121.
Kaur S, Van Bergen NJ, Gold WA, Eggers S, Lunke S, White SM, et al. Whole exome sequencing reveals a de novo missense variant in EEF1A2 in a Rett syndrome-like patient. Clin Case Rep. 2019;7:2476–82. https://doi.org/10.1002/ccr3.2511.
Zacher P, Mayer T, Brandhoff F, Bartolomaeus T, Le Duc D, Finzel M, et al. The genetic landscape of intellectual disability and epilepsy in adults and the elderly: a systematic genetic work-up of 150 individuals. Genet Med J Am Coll Med Genet. 2021;23:1492–7. https://doi.org/10.1038/s41436-021-01153-6.
Lopes F, Barbosa M, Ameur A, Soares G, de Sà J, Dias AI, et al. Identification of novel genetic causes of Rett syndrome-like phenotypes. J Med Genet. 2016;53:190–9. https://doi.org/10.1136/jmedgenet-2015-103568.
Lam WWK, Millichap JJ, Soares DC, Chin R, McLellan A, FitzPatrick DR, et al. Novel de novo EEF1A2 missense mutations causing epilepsy and intellectual disability. Mol Genet Genom Med. 2016;4:465–74. https://doi.org/10.1002/mgg3.219.
de Kovel CGF, Brilstra EH, van Kempen MJA, Van’t Slot R, Nijman IJ, Afawi Z, et al. Targeted sequencing of 351 candidate genes for epileptic encephalopathy in a large cohort of patients. Mol Genet Genom Med. 2016;4:568–80. https://doi.org/10.1002/mgg3.235.
Nakajima J, Okamoto N, Tohyama J, Kato M, Arai H, Funahashi O, et al. De novo EEF1A2 mutations in patients with characteristic facial features, intellectual disability, autistic behaviors and epilepsy. Clin Genet. 2015;87:356–61. https://doi.org/10.1111/cge.12394.
Ostrander BEP, Butterfield RJ, Pedersen BS, Farrell AJ, Layer RM, Ward A, et al. Whole-genome analysis for effective clinical diagnosis and gene discovery in early infantile epileptic encephalopathy. NPJ Genom Med. 2018;3:22. https://doi.org/10.1038/s41525-018-0061-8.
De Rinaldis M, Giorda R, Trabacca A. Mild epileptic phenotype associates with de novo eef1a2 mutation: Case report and review. Brain Dev. 2020;42:77–82. https://doi.org/10.1016/j.braindev.2019.08.001.
Lance EI, Kronenbuerger M, Cohen JS, Furmanski O, Singer HS, Fatemi A. Successful treatment of choreo-athetotic movements in a patient with an EEF1A2 gene variant. SAGE Open Med Case Rep. 2018;6:2050313X18807622. https://doi.org/10.1177/2050313X18807622.
O’Roak BJ, Stessman HA, Boyle EA, Witherspoon KT, Martin B, Lee C, et al. Recurrent de novo mutations implicate novel genes underlying simplex autism risk. Nat Commun. 2014;5:5595. https://doi.org/10.1038/ncomms6595.
Helbig KL, Farwell Hagman KD, Shinde DN, Mroske C, Powis Z, Li S, et al. Diagnostic exome sequencing provides a molecular diagnosis for a significant proportion of patients with epilepsy. Genet Med. 2016;18:898–905. https://doi.org/10.1038/gim.2015.186.
Vogt LM, Lorenzo M, Prendergast B, Jobling D, Gill R. PJ. EEF1A2 pathogenic variant presenting in an infant with failure to thrive and frequent apneas requiring respiratory support. Am J Med Genet A. 2022;188:3106–9. https://doi.org/10.1002/ajmg.a.62932.
Epi25 Collaborative. Electronic address: jm4279@cumc.columbia.edu; Epi25 Collaborative. Sub-genic intolerance, ClinVar, and the epilepsies: A whole-exome sequencing study of 29,165 individuals. Am J Hum Genet. 2021;108:965–82. https://doi.org/10.1016/j.ajhg.2021.04.009.
McNicholas S, Potterton E, Wilson KS, Noble MEM. Presenting your structures: the CCP4mg molecular-graphics software. Acta Cryst 2011;D67:386–94.
Ittisoponpisan S, Islam SA, Khanna T, Alhuzimi E, David A, Sternberg MJE. Can Predicted Protein 3D Structures Provide Reliable Insights into whether Missense Variants Are Disease Associated? J Mol Biol 2019;431:2197–212. https://doi.org/10.1016/j.jmb.2019.04.009.
Van Durme J, Delgado J, Stricher F, Serrano L, Schymkowitz J, Rousseau F. A graphical interface for the FoldX forcefield. Bioinformatics. 2011;27:1711–2.
Acknowledgements
We want to acknowledge AnDDI-Rares and the families for their participation. In addition, the collaboration in this study were facilitated by ERN ITHACA, one of the 24 European Reference Networks (ERNs) approved by the ERN Board of Member States, cofounded by European Commission. [EU Framework Partnership Agreement ID: 3HP-HP-FPA ERN-01-2016/739516].
Funding
The aims of this study contribute to the Solve-RD project, which has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement number 779257.
Author information
Authors and Affiliations
Contributions
Conceptualization: AP, LR, CA, AV. Formal Analysis: AP. Investigation: AP, FA, LR, MCK, SB, JL, DT, SB, MCK, AV, RT, CBA, MG, BG, SP, AO, CZ, TS, MW, LF, MS, PS, IB, DK, MI, MS. Supervision: LR, AV, CA. Visualization: AP, CA, CBN. Writing - Original Draft: AP, LR, CA, CBN. Writing - Review & Editing: AP, LR, CA, CBN, AV, FM, CC, RF, JP, SJ, CF, LW, JF, AV, ASDP, TH, JM, SAL, WL, RJ, MK, DG, AG, EDB, JL, DT, SB, MCK, AV, RT, CBA, MG, BG, SP, DK.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
Ethical approval was not required because it was a retrospective observational study and patients have already signed consent for genetics analyses for diagnosis. Every patient has been anonymized by the clinician before collecting data (a number has been assigned to each patient).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: In the original version of this article, the given and family names of Samuel Groeschel were incorrectly structured. The name was displayed correctly in all versions at the time of publication. The authors, Adam Ostendorf, Christiane Zweier, Thomas Smol, Marjolaine Willems, Laurence Faivre, Marcello Scala, Pasquale Striano, Irene Bagnasco, Daniel Koboldt, Maria Iascone, Manon Suerink were missing from the author list. Moreover, the author contribution texts have been updated in accordance with addition of authors.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Paulet, A., Bennett-Ness, C., Ageorges, F. et al. Expansion of the neurodevelopmental phenotype of individuals with EEF1A2 variants and genotype-phenotype study. Eur J Hum Genet (2024). https://doi.org/10.1038/s41431-024-01560-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41431-024-01560-8