Abstract
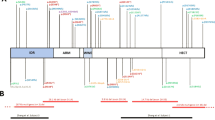
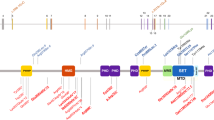
Mutations in ADNP result in Helsmoortel–Van der Aa syndrome. Here, we describe the first de novo intronic deletion, affecting the splice-acceptor site of the first coding ADNP exon in a five-year-old girl with developmental delay and autism. Whereas exome sequencing failed to detect the non-coding deletion, genome-wide CpG methylation analysis revealed an episignature suggestive of a Helsmoortel–Van der Aa syndrome diagnosis. This diagnosis was further supported by PhenoScore, a novel facial recognition software package. Subsequent whole-genome sequencing resolved the three-base pair ADNP deletion c.[-5-1_-4del] with transcriptome sequencing showing this deletion leads to skipping of exon 4. An N-terminal truncated protein could not be detected in transfection experiments with a mutant expression vector in HEK293T cells, strongly suggesting this is a first confirmed diagnosis exclusively due to haploinsufficiency of the ADNP gene. Pathway analysis of the methylome indicated differentially methylated genes involved in brain development, the cytoskeleton, locomotion, behavior, and muscle development. Along the same line, transcriptome analysis identified most of the differentially expressed genes as upregulated, in line with the hypomethylated CpG episignature and confirmed the involvement of the cytoskeleton and muscle development pathways that are also affected in patient cell lines and animal models. In conclusion, this novel mutation for the first time demonstrates that Helsmoortel–Van der Aa syndrome can be caused by a loss-of-function mutation. Moreover, our study elegantly illustrates the use of EpiSignatures, WGS and Phenoscore as novel complementary diagnostic tools in case a of negative WES result.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The datasets generated during this study are available from the corresponding author upon argumented request.
References
Pinhasov A, Mandel S, Torchinsky A, Giladi E, Pittel Z, Goldsweig AM, et al. Activity-dependent neuroprotective protein: a novel gene essential for brain formation. Brain Res Dev Brain Res. 2003;144:83–90.
Mandel S, Rechavi G, Gozes I. Activity-dependent neuroprotective protein (ADNP) differentially interacts with chromatin to regulate genes essential for embryogenesis. Dev Biol. 2007;303:814–24.
Mandel S, Gozes I. Activity-dependent neuroprotective protein constitutes a novel element in the SWI/SNF chromatin remodeling complex. J Biol Chem. 2007;282:34448–56.
Ostapcuk V, Mohn F, Carl SH, Basters A, Hess D, Iesmantavicius V, et al. Activity-dependent neuroprotective protein recruits HP1 and CHD4 to control lineage-specifying genes. Nature. 2018;557:739–43.
D’Incal CP, Van Rossem KE, De Man K, Konings A, Van Dijck A, Rizzuti L, et al. Chromatin remodeler Activity-Dependent Neuroprotective Protein (ADNP) contributes to syndromic autism. Clin Epigenetics. 2023;15:45.
Van Dijck A, Vulto-van Silfhout AT, Cappuyns E, van der Werf IM, Mancini GM, Tzschach A, et al. Clinical presentation of a complex neurodevelopmental disorder caused by mutations in ADNP. Biol Psychiatry. 2019;85:287–97.
Helsmoortel C, Vulto-van Silfhout AT, Coe BP, Vandeweyer G, Rooms L, van den Ende J, et al. A SWI/SNF-related autism syndrome caused by de novo mutations in ADNP. Nat Genet. 2014;46:380–4.
Vandeweyer G, Helsmoortel C, Van Dijck A, Vulto-van Silfhout AT, Coe BP, Bernier R, et al. The transcriptional regulator ADNP links the BAF (SWI/SNF) complexes with autism. Am J Med Genet C Semin Med Genet. 2014;166C:315–26.
Bend EG, Aref-Eshghi E, Everman DB, Rogers RC, Cathey SS, Prijoles EJ, et al. Gene domain-specific DNA methylation episignatures highlight distinct molecular entities of ADNP syndrome. Clin Epigenetics. 2019;11:64.
Breen MS, Garg P, Tang L, Mendonca D, Levy T, Barbosa M, et al. Episignatures Stratifying Helsmoortel-Van Der Aa Syndrome Show Modest Correlation with Phenotype. Am J Hum Genet. 2020;107:555–63.
Dingemans AJM, Hinne M, Truijen KMG, Goltstein L, van Reeuwijk J, de Leeuw N, et al. PhenoScore quantifies phenotypic variation for rare genetic diseases by combining facial analysis with other clinical features using a machine-learning framework. Nat Genet. 2023;55:1598–607.
D’Incal CP, Cappuyns E, Choukri K, Szrama K, De Man K, Van der Aa N et al. In Search of the Hidden Protein: Optimization of Detection Strategies for autism-associated Activity-Dependent Neuroprotective Protein (ADNP) mutants. Res Sq. 2022. https://doi.org/10.21203/rs.3.rs-1954095/v1.
Georget M, Lejeune E, Buratti J, Servant E, le Guern E, Heron D, et al. Loss of function of ADNP by an intragenic inversion. Eur J Hum Genet. 2023;31:967–70.
D’Incal CP, Kooy RF. ADNP in reverse gear. Eur J Hum Genet. 2023;31:849–50.
Akhmedov M, Martinelli A, Geiger R, Kwee I. Omics Playground: a comprehensive self-service platform for visualization, analytics and exploration of Big Omics Data. NAR Genom Bioinform. 2020;2:lqz019.
Zhou W, Triche TJ, Laird PW, Shen H. SeSAMe: reducing artifactual detection of DNA methylation by Infinium BeadChips in genomic deletions. Nucleic Acids Res. 2018;46:e123.
Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523.
Gozes I, Van Dijck A, Hacohen-Kleiman G, Grigg I, Karmon G, Giladi E, et al. Premature primary tooth eruption in cognitive/motor-delayed ADNP-mutated children. Transl Psychiatry. 2017;7:e1043.
Anna A, Monika G. Splicing mutations in human genetic disorders: examples, detection, and confirmation. J Appl Genet. 2018;59:253–68.
Huynh M-T, Boudry-Labis E, Massard A, Thuillier C, Delobel B, Duban-Bedu B, et al. A heterozygous microdeletion of 20q13.13 encompassing ADNP gene in a child with Helsmoortel-van der Aa syndrome. Eur J Hum Genet. 2018;26:1497–501.
Cappuyns E, Huyghebaert J, Vandeweyer G, Kooy RF. Mutations in ADNP affect expression and subcellular localization of the protein. Cell Cycle. 2018;17:1068–75.
Ivashko-Pachima Y, Sayas CL, Malishkevich A, Gozes I. ADNP/NAP dramatically increase microtubule end-binding protein-Tau interaction: a novel avenue for protection against tauopathy. Mol Psychiatry. 2017;22:1335–44.
Kapitansky O, Karmon G, Sragovich S, Hadar A, Shahoha M, Jaljuli I et al. Single cell ADNP predictive of human muscle disorders: mouse knockdown results in muscle wasting. Cells. 2020;9. https://doi.org/10.3390/cells9102320.
Acknowledgements
The authors would like to thank the family for participating in this study.
Funding
RFK acknowledges the support of the Research Fund of the University of Antwerp OEC-Methusalem grant “GENOMED”. This work was in part financed by grants from the ERA-NET NEURON “ADNPinMED”. This article is also based upon work from COST Action International Nucleome Consortium (INC) CA18127, supported by COST (European Cooperation in Science and Technology) ascribed to WVB.
Author information
Authors and Affiliations
Contributions
CPD performed the experiments, conceptualized the experimental design, performed data analysis, wrote the manuscript text, and prepared all the figures. DJA provided the bioinformatic analysis of the methylation array. EE conducted all the gene expression assays using RT-PCR under supervision of CD, JJVDS and MA performed the WES, WGS, and EpiSign at early observations of the patient, AJMD and BBADV provided analysis of the ADNP female using PhenoScore. LM provided the bioinformatic analysis of the RNA sequencing. WVB and RFK reviewed and edited the manuscript. All authors reviewed and approved the final version of manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
Written informed consent for publication of individual details and images was obtained in the context of the Helsmoortel–Van der Aa syndrome from the parents. The protocol was approved by the Ethics Committee of the Antwerp University Hospital, Antwerp, Belgium.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
D’Incal, C.P., Annear, D.J., Elinck, E. et al. Loss-of-function of activity-dependent neuroprotective protein (ADNP) by a splice-acceptor site mutation causes Helsmoortel–Van der Aa syndrome. Eur J Hum Genet (2024). https://doi.org/10.1038/s41431-024-01556-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41431-024-01556-4