Abstract
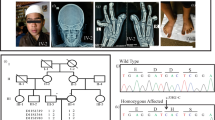
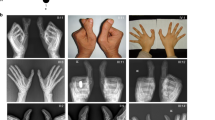
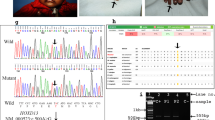
Polydactyly is the most common limb malformation that occurs in 1.6–10.6 per one thousand live births, with incidence varying with ancestry. The underlying gene has been identified for many of the ~100 syndromes that include polydactyly. While for the more common form, nonsydromic polydactyly, eleven candidate genes have been reported. We investigated the underlying genetic cause of autosomal recessive nonsyndromic postaxial polydactyly in four consanguineous Pakistani families. Some family members with postaxial polydactyly also present with syndactyly, camptodactyly, or clinodactyly. Analysis of the exome sequence data revealed two novel homozygous frameshift deletions in EFCAB7: [c.830delG;p.(Gly277Valfs*5)]; in three families and [c.1350_1351delGA;p.(Asn451Phefs*2)] in one family. Sanger sequencing confirmed that these variants segregated with postaxial polydactyly, i.e., family members with postaxial polydactyly were found to be homozygous while unaffected members were heterozygous or wild type. EFCAB7 displays expressions in the skeletal muscle and on the cellular level in cilia. IQCE-EFCAB7 and EVC-EVC2 are part of the heterotetramer EvC complex, which is a positive regulator of the Hedgehog (Hh) pathway, that plays a key role in limb formation. Depletion of either EFCAB7 or IQCE inhibits induction of Gli1, a direct Hh target gene. Variants in IQCE and GLI1 have been shown to cause nonsyndromic postaxial polydactyly, while variants in EVC and EVC2 underlie Ellis van Creveld and Weyers syndromes, which include postaxial polydactyly as a phenotype. This is the first report of the involvement of EFCAB7 in human disease etiology.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
The identified variants have been submitted to the ClinVar database [Accession numbers: SCV003804323 and SCV003804374].
References
Ahmad Z, Liaqat R, Palander O, Bilal M, Zeb S, Ahmad F, et al. Genetic overview of postaxial polydactyly: Updated classification. Clin Genet. 2023;103:3–15.
Umair M, Ahmad F, Bilal M, Ahmad W, Alfadhel M. Clinical genetics of polydactyly: An updated review. Front Genet. 2018;9:447.
Pompe van Meerdervoort HF. Congenital musculoskeletal malformation in South Africa Blakcs-a study of incidence. S Afr Med J. 1976;50:1853–5.
Woolf CM, Myrianthopoulos NC. Polydactyly in American negroes and whites. American. J Hum Genet. 1973;25:397.
Kondoh S, Sugawara H, Harada N, Matsumoto N, Ohashi H, Sato M, et al. A novel gene is disrupted at a 14q13 breakpoint of t(2;14) in a patient with mirror-image polydactyly of hands and feet. J Hum Genet. 2002;47:136–9.
Klopocki E, Kähler C, Foulds N, Shah H, Joseph B, Vogel H, et al. Deletions in PITX1 cause a spectrum of lower-limb malformations including mirror-image polydactyly. Eur J Hum Genet. 2012;20:705–8.
Kalsoom UE, Klopocki E, Wasif N, Tariq M, Khan S, Hecht J, et al. Whole exome sequencing identified a novel zinc-finger gene ZNF141 associated with autosomal recessive postaxial polydactyly type A. J Med Genet. 2013;50:47–53.
Bakar A, Ullah A, Bibi N, Khan H, Rahman AU, Ahmad W, et al. A novel homozygous variant in the GLI1 underlies postaxial polydactyly in a large consanguineous family with intra familial variable phenotypes. Eur J Med Genet. 2022;65:104599.
Umair M, Shah K, Alhaddad B, Haack TB, Graf E, Strom TM, et al. Exome sequencing revealed a splice site variant in the IQCE gene underlying post-axial polydactyly type A restricted to lower limb. Eur J Hum Genet. 2017;25:960–5.
Ullah I, Kakar N, Schrauwen I, Hussain S, Chakchouk I, Liaqat K, et al. Variants in KIAA0825 underlie autosomal recessive postaxial polydactyly. Hum Genet. 2019;138:593–600.
Umair M, Wasif N, Albalawi AM, Ramzan K, Alfadhel M, Ahmad W, et al. Exome sequencing revealed a novel loss‐of‐function variant in the GLI3 transcriptional activator 2 domain underlies nonsyndromic postaxial polydactyly. Mol Genet Genom Med. 2019;7:e00627.
Schrauwen I, Giese AP, Aziz A, Lafont DT, Chakchouk I, Santos-Cortez RLP, et al. FAM92A underlies nonsyndromic postaxial polydactyly in humans and an abnormal limb and digit skeletal phenotype in mice. J Bone Min Res. 2019;34:375–86.
Umair M, Palander O, Bilal M, Almuzzaini B, Alam Q, Ahmad F, et al. Biallelic variant in DACH1, encoding Dachshund Homolog 1, defines a novel candidate locus for recessive postaxial polydactyly type A. Genomics 2021;113:2495–502.
Pusapati GV, Hughes CE, Dorn KV, Zhang D, Sugianto P, Aravind L, et al. EFCAB7 and IQCE regulate hedgehog signaling by tethering the EVC-EVC2 complex to the base of primary cilia. Dev Cell. 2014;28:483–96.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;2009:1754–60.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome AnalysisToolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303.
Yang H, Wang K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat Protoc. 2015;10:1556–66.
Kircher M, Witten DM, Jain P, O’roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014 Mar;46:310–5.
Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020;581:434–43.
Kowalski MH, Qian H, Hou Z, Rosen JD, Tapia AL, Shan Y, et al. Use of >100,000 NHLBI Trans-Omics for PrecisionMedicine (TOPMed) Consortium whole genome sequences improves imputation quality anddetection of rare variant associations in admixed African and Hispanic/Latino populations. PLOS Genet. 2019;15:e1008500.
Scott EM, Halees A, Itan Y, Spencer EG, He Y, Azab MA, et al. Characterization of Greater Middle Eastern genetic variation for enhanced disease gene discovery. Nat Genet. 2016;48:1071–6.
Lyles CR, Lunn MR, Obedin-Maliver J, Bibbins-Domingo K. The new era of precision population health: insights for the All of Us Research Program and beyond. J Transl Med. 2018;16:1–4.
Khan H, Ahmed S, Nawaz S, Ahmad W, Rafiq MA. Greig cephalopolysyndactyly syndrome: phenotypic variability associated with variants in two different domains of GLI3. Klin Padiatr. 2021;233:53–8.
Zhang Z, Xin D, Wang P, Zhou L, Hu L, Kong X, et al. Noisy splicing, more than expression regulation, explains why some exons are subject to nonsense-mediated mRNA decay. BMC Biol. 2009;7:1–3.
Dorn KV, Hughes CE, Rohatgi R. A Smoothened-Evc2 complex transduces the Hedgehog signal at primary cilia. Dev Cell. 2012;23:823–35.
Gigante ED, Caspary T. Signaling in the primary cilium through the lens of the Hedgehog pathway. Wiley Interdiscip Rev: Dev Biol. 2020;9:e377.
Speir ML, Bhaduri A, Markov NS, Moreno P, Nowakowski TJ, Papatheodorou I, et al. UCSC Cell Browser: visualize your single-cell data. Bioinformatics 2021;37:4578–80.
Muñoz-Fuentes V, Cacheiro P, Meehan TF, Aguilar-Pimentel JA, Brown SD, Flenniken AM, et al. The International Mouse Phenotyping Consortium (IMPC): a functional catalogue of the mammalian genome that informs conservation. Conserv Genet. 2018;19:995–1005.
Breslow DK, Hoogendoorn S, Kopp AR, Morgens DW, Vu BK, Kennedy MC, et al. A CRISPR-based screen for Hedgehog signaling provides insights into ciliary function and ciliopathies. Nat Genet. 2018;50:460–71.
Umair M, Seidel H, Ahmed I, Ullah A, Haack TB, Alhaddad B, et al. Ellis–van Creveld syndrome and profound deafness resulted by sequence variants in the EVC/EVC2 and TMC1 genes. J Genet. 2017;96:1005–14.
Hussain S, Nawaz S, Khan H, Acharya A, Schrauwen I, Ahmad W, et al. A splice site variant in TCTN3 underlies an atypical form of orofaciodigital syndrome IV. Ann Hum Genet. 2022;86:291–6.
Shamseldin HE, Shaheen R, Ewida N, Bubshait DK, Alkuraya H, Almardawi E, et al. The morbid genome of ciliopathies: an update. Genet Med. 2020;22:1051–60.
Nguyen TQ, Doan NM, Trinh HT, Mizuguchi M. Novel mutation in EFCAB7 alters expression and interaction of Ellis–van Creveld ciliary proteins. Congenit Anom. 2019;59:49–50.
Acknowledgements
We are highly obliged to members of the families for their cooperation and participation in this study. Muhammad Bilal, Hammal Khan and Muhammad Javed Khan were supported by Indigenous PhD and IRSIP fellowships from Higher Education Commission (HEC), Islamabad, Pakistan.
Funding
This research was funded by Higher Education Commission of Pakistan, Pediatric Genomics Discovery Program, and the Department of Neurology, Columbia University Medical Center.
Author information
Authors and Affiliations
Contributions
MB, HK, and MJK performed the laboratory experiments, analyzed the genomic data, and drafted the manuscript. IS, KL, AA, TB, N, HA, SA, KU, MSH, and AU collected samples and analyzed clinical and genomic data. WA and SML designed the project, analyzed the data, edited the manuscript, and provided funds for the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interest.
Ethical approval
Written informed consent for presentation and publication of this study was obtained from all the participating members.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bilal, M., Khan, H., Khan, M.J. et al. Variants in EFCAB7 underlie nonsyndromic postaxial polydactyly. Eur J Hum Genet 31, 1270–1274 (2023). https://doi.org/10.1038/s41431-023-01450-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-023-01450-5
This article is cited by
-
2023 in the European Journal of Human Genetics
European Journal of Human Genetics (2024)
-
Deep phenotyping and population-level data can help resolve genomic variants
European Journal of Human Genetics (2023)