Abstract
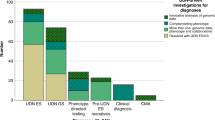
The Korean Genetic Diagnosis Program for Rare Disease (KGDP) enrolled 1890 patients with rare diseases between March 2017 and October 2022. Children and adolescents accounted for the majority of the patients, and systemic disease was the most common presenting symptom. The exome-based virtual disease-specific multigene panel was the most frequently used analytical method, with an overall diagnostic yield of 33.3%. A total of 629 positive cases were diagnosed, involving 297 genes. All 297 genes identified in these cases were confirmed to be known genes listed in the OMIM database. The nationwide KGDP network and its cooperation with the Korean Undiagnosed Diseases Program (KUDP) provide a more comprehensive genetic analysis of undiagnosed cases. The partnership between the KGDP and KUDP has the potential to improve the diagnosis and treatment options for patients. In conclusion, KGDP serves as the primary access point or gateway to KUDP.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Hayashi S, Umeda T. 35 years of Japanese policy on rare diseases. Lancet 2008;372:889–90.
Puiu M, Dan D. Rare diseases, from European resolutions and recommendations to actual measures and strategies. Maedica. 2010;5:128–31.
Valdez R, Ouyang L, Bolen J. Public health and rare diseases: oxymoron no more. Prev Chronic Dis. 2016;13:E05.
Marwaha S, Knowles JW, Ashley EA. A guide for the diagnosis of rare and undiagnosed disease: beyond the exome. Genome Med. 2022;14:23.
The Lancet Diabetes Endocrinology. Spotlight on rare diseases. Lancet Diabetes Endocrinol. 2019;7:75.
Navarrete-Opazo AA, Singh M, Tisdale A, Cutillo CM, Garrison SR. Can you hear us now? The impact of health-care utilization by rare disease patients in the United States. Genet Med. 2021;23:2194–201.
Adachi T, Kawamura K, Furusawa Y, Nishizaki Y, Imanishi N, Umehara S, et al. Japan’s initiative on rare and undiagnosed diseases (IRUD): towards an end to the diagnostic odyssey. Eur J Hum Genet. 2017;25:1025–8.
Beaulieu CL, Majewski J, Schwartzentruber J, Samuels ME, Fernandez BA, Bernier FP, et al. FORGE Canada Consortium: outcomes of a 2-year national rare-disease gene-discovery project. Am J Hum Genet. 2014;94:809–17.
Kim SY, Lim BC, Lee JS, Kim WJ, Kim H, Ko JM, et al. The Korean undiagnosed diseases program: lessons from a one-year pilot project. Orphanet J Rare Dis. 2019;14:68.
Lochmuller H, Torrent IFJ, Le Cam Y, Jonker AH, Lau LP, Baynam G, et al. The International Rare Diseases Research Consortium: policies and guidelines to maximize impact. Eur J Hum Genet. 2017;25:1293–302.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Kim MJ, Lee S, Yun H, Cho SI, Kim B, Lee JS, et al. Consistent count region-copy number variation (CCR-CNV): an expandable and robust tool for clinical diagnosis of copy number variation at the exon level using next-generation sequencing data. Genet Med. 2022;24:663–72.
Kim SY, Lee S, Woo H, Han J, Ko YJ, Shim Y, et al. The Korean undiagnosed diseases program phase I: expansion of the nationwide network and the development of long-term infrastructure. Orphanet J Rare Dis. 2022;17:372.
Keicho N, Hijikata M, Morimoto K, Homma S, Taguchi Y, Azuma A, et al. Primary ciliary dyskinesia caused by a large homozygous deletion including exons 1-4 of DRC1 in Japanese patients with recurrent sinopulmonary infection. Mol Genet Genom Med. 2020;8:e1033.
Morimoto K, Hijikata M, Zariwala MA, Nykamp K, Inaba A, Guo TC, et al. Recurring large deletion in DRC1 (CCDC164) identified as causing primary ciliary dyskinesia in two Asian patients. Mol Genet Genom Med. 2019;7:e838.
Takeuchi K, Xu Y, Kitano M, Chiyonobu K, Abo M, Ikegami K, et al. Copy number variation in DRC1 is the major cause of primary ciliary dyskinesia in the Japanese population. Mol Genet Genom Med. 2020;8:e1137.
Kim MJ, Kim S, Chae SW, Lee S, Yoon JG, Kim B, et al. Prevalence and founder effect of DRC1 exon 1-4 deletion in Korean patients with primary ciliary dyskinesia. J Hum Genet. 2023;68:369–74.
Srivastava S, Love-Nichols JA, Dies KA, Ledbetter DH, Martin CL, Chung WK, et al. Meta-analysis and multidisciplinary consensus statement: exome sequencing is a first-tier clinical diagnostic test for individuals with neurodevelopmental disorders. Genet Med. 2019;21:2413–21.
Shickh S, Gutierrez Salazar M, Zakoor KR, Lazaro C, Gu J, Goltz J, et al. Exome and genome sequencing in adults with undiagnosed disease: a prospective cohort study. J Med Genet. 2021;58:275–83.
Han SR, Lee YA, Shin CH, Yang SW, Lim BC, Cho TJ, et al. Clinical and molecular characteristics of GNAS inactivation disorders observed in 18 Korean patients. Exp Clin Endocrinol Diabetes. 2021;129:118–25.
Kim HY, Shin CH, Lee YA, Shin CH, Kim GH, Ko JM. Deciphering epigenetic backgrounds in a Korean cohort with Beckwith-Wiedemann syndrome. Ann Lab Med. 2022;42:668–77.
Kim B, Kim MJ, Hur K, Jo SJ, Ko JM, Park SS, et al. Clinical and genetic profiling of nevoid basal cell carcinoma syndrome in Korean patients by whole-exome sequencing. Sci Rep. 2021;11:1163.
Funding
This study was supported by a grant (6500-6534-306) from the Korea Disease Control and Prevention Agency from 2021. Prior to this, from 2020 and earlier, our research was funded by a grant (4860-306-320) from the Korea Centers for Disease Control and Prevention.
Author information
Authors and Affiliations
Contributions
Concept and design: MJK and MWS. Acquisition, analysis, or interpretation of data: all authors. Drafting of the manuscript: MJK and MWS. Statistical analysis: MJK. Obtained funding: MWS. Review of the manuscript: all authors. Supervision: MWS.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
All study procedures were approved by the Institutional Review Board of SNUH (IRB number: 2212-114-1389), and the study adhered to the Declaration of Helsinki for biomedical research involving human subjects.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kim, M.J., Kim, B., Lee, H. et al. The Korean Genetic Diagnosis Program for Rare Disease Phase II: outcomes of a 6-year national project. Eur J Hum Genet 31, 1147–1153 (2023). https://doi.org/10.1038/s41431-023-01415-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-023-01415-8
This article is cited by
-
Expanding what we know about rare genetic diseases
European Journal of Human Genetics (2023)