Abstract
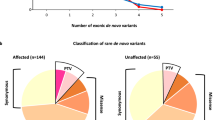
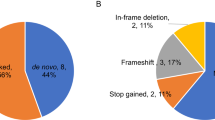
De novo variants (DNVs) analysis has proven to be a powerful approach to gene discovery in Autism Spectrum Disorder (ASD), which has not yet been shown in a Brazilian ASD cohort. The relevance of inherited rare variants has also been suggested, particularly in oligogenic models. We hypothesized that three-generation analyses of DNVs could provide new insights into the relevance of de novo and inherited variants across generations. To accomplish this goal, we performed whole-exome sequencing of 33 septet families composed of probands, parents, and grandparents (n = 231 individuals) and compared DNV rates (DNVr) between generations and those from two control cohorts. The DNVr in the probands (DNVr = 1.16) was marginally higher than in parents (DNVr = 0.60; p = 0.054), and in controls (DNVr = 0.68; p = 0.035, congenital heart disorder and DNVr = 0.70; p = 0.047, unaffected ASD siblings from Simons Simplex Collection). Moreover, most of the DNVs were found to have paternal origin in both generations (84.6%). Finally, we observed that 40% (6/15) of the DNVs in parents transmitted for probands are in ASD or ASD candidate genes, representing recently emerged risk variants to ASD in their families and suggest ZNF536, MSL2 and HDAC9 as ASD candidate genes. We did not observe an enrichment of risk variants nor sex bias of transmitted variants in the three generations, that can be due to sample size. These results further reinforce the relevance of de novo variants in ASD.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
Data availability
Data generated as part of this study are available from the corresponding author on reasonable request. All de novo variants described in this work were submitted to the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/), accession numbers SCV003914734 - SCV003914804.
References
Association AP, Kernberg. Diagnostic and Statistical Manual of Mental Disorders. 5th edition. Washington, D.C.: American Psychiatric Publishing; 2013.
Lord C, Brugha TS, Charman T, Cusack J, Dumas G, Frazier T, et al. Autism spectrum disorder. Nat Rev Dis Primers. 2020;6:1–23.
Woodbury-Smith M, Scherer SW. Progress in the genetics of autism spectrum disorder. Dev Med Child Neurol. 2018;60:445–51.
Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–21.
Pinto D, Delaby E, Merico D, Barbosa M, Merikangas A, Klei L, et al. Convergence of Genes and Cellular Pathways Dysregulated in Autism Spectrum Disorders. Am J Hum Genet. 2014;94:677–94.
Yuen RK C, Merico D, Bookman ML, Howe J, Thiruvahindrapuram B, Patel RV, et al. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat Neurosci. 2017;20:602–11.
Zarrei M, Burton CL, Engchuan W, Young EJ, Higginbotham EJ, MacDonald JR, et al. A large data resource of genomic copy number variation across neurodevelopmental disorders. NPJ Genom Med. 2019;4:26.
Satterstrom FK, Kosmicki JA, Wang J, Breen MS, De Rubeis S, An JY, et al. Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell. 2020;180:568–84.e23.
Turner TN, Coe BP, Dickel DE, Hoekzema K, Nelson BJ, Zody MC, et al. Genomic patterns of De Novo mutation in simplex autism. Cell. 2017;171:710–22.e12.
da Silva Montenegro EM, Costa CS, Campos G, Scliar M, de Almeida TF, Zachi EC, et al. Meta-analyses support previous and novel autism candidate genes: Outcomes of an unexplored Brazilian cohort. Autism Res. 2020;13:199–206.
Ruzzo EK, Pérez-Cano L, Jung JY, Wang LK, Kashef-Haghighi D, Hartl C, et al. Inherited and De Novo genetic risk for autism impacts shared networks. Cell. 2019;178:850–66.e26.
Wilfert AB, Turner TN, Murali SC, Hsieh P, Sulovari A, Wang T, et al. Recent ultra-rare inherited variants implicate new autism candidate risk genes. Nat Genet. 2021;53:1125–34.
Krumm N, Turner TN, Baker C, Vives L, Mohajeri K, Witherspoon K, et al. Excess of rare, inherited truncating mutations in autism. Nat Genet. 2015;47:582–8.
Yuen RKC, Merico D, Cao H, Pellecchia G, Alipanahi B, Thiruvahindrapuram B, et al. Genome-wide characteristics of de novo mutations in autism. NPJ Genom Med. 2016;1:160271–10.
Jónsson H, Sulem P, Kehr B, Kristmundsdottir S, Zink F, Hjartarson E, et al. Parental influence on human germline de novo mutations in 1,548 trios from Iceland. Nature. 2017;549:519–22.
Gao Z, Moorjani P, Sasani TA, Pedersen BS, Quinlan AR, Jorde LB, et al. Overlooked roles of DNA damage and maternal age in generating human germline mutations. Proc Natl Acad Sci USA. 2019;116:9491–500.
Kong A, Frigge ML, Masson G, Besenbacher S, Sulem P, Magnusson G, et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488:471–5.
Homsy J, Zaidi S, Shen Y, Ware JS, Samocha KE, Karczewski KJ, et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015;350:1262–6.
Sifrim A, Hitz MP, Wilsdon A, Breckpot J, Turki SHA, Thienpont B, et al. Distinct genetic architectures for syndromic and nonsyndromic congenital heart defects identified by exome sequencing. Nat Genet. 2016;48:1060–5.
Miles JH, Takahashi TN, Bagby S, Sahota PK, Vaslow DF, Wang CH, et al. Essential versus complex autism: Definition of fundamental prognostic subtypes. Am J Med Genet Part A. 2005;135A:171–80.
Ramu A, Noordam MJ, Schwartz RS, Wuster A, Hurles ME, Cartwright RA, et al. DeNovoGear: de novo indel and point mutation discovery and phasing. Nat Methods. 2013;10:985–7.
Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6.
Du Y, Li Z, Liu Z, Zhang N, Wang R, Li F, et al. Nonrandom occurrence of multiple de novo coding variants in a proband indicates the existence of an oligogenic model in autism. Genet Med. 2020;22:170–80.
Wang T, Zhao PA, Eichler EE. Rare variants and the oligogenic architecture of autism. Trends Genet. 2022;38:895–903.
Pizzo L, Jensen M, Polyak A, Rosenfeld JA, Mannik K, Krishnan A, et al. Rare variants in the genetic background modulate cognitive and developmental phenotypes in individuals carrying disease-associated variants. Genet Med. 2019;21:816–25.
Hiatt SM, Thompson ML, Prokop JW, Lawlor JMJ, Gray DE, Bebin EM, et al. Deleterious Variation in BRSK2 Associates with a Neurodevelopmental Disorder. Am J Hum Genet. 2019;104:701–8.
Qin Z, Ren F, Xu X, Ren Y, Li H, Wang Y, et al. ZNF536, a novel zinc finger protein specifically expressed in the brain, negatively regulates neuron differentiation by repressing retinoic acid-induced gene transcription. Mol Cell Biol. 2009;29:3633–43.
Thyme SB, Pieper LM, Li EH, Pandey S, Wang Y, Morris NS, et al. Phenotypic landscape of schizophrenia-associated genes defines candidates and their shared functions. Cell. 2019;177:478–91.e20.
Wu L, Zee BM, Wang Y, Garcia BA, Dou Y. The RING finger protein MSL2 in the MOF complex is an E3 ubiquitin ligase for H2B K34 and is involved in crosstalk with H3 K4 and K79 methylation. Mol Cell. 2011;43:132–44.
Volmar CH, Wahlestedt C. Histone deacetylases (HDACs) and brain function. Neuroepigenetics. 2015;1:20–7.
Lang B, Alrahbeni TMA, Clair DS, Blackwood DH. HDAC9 is implicated in schizophrenia and expressed specifically in post-mitotic neurons but not in adult neural stem cells. Am J Stem Cells. 2012;1:31–41.
Krumm N, Turner TN, Baker C, Vives L, Mohajeri K, Witherspoon K, et al. Excess of rare, inherited truncating mutations in autism. Nat Genet. 2015;47:582–8.
Krumm N, O’Roak BJ, Karakoc E, Mohajeri K, Nelson B, Vives L, et al. Transmission disequilibrium of small CNVs in simplex autism. Am J Hum Genet. 2013;93:595–606.
Brandler WM, Antaki D, Gujral M, Kleiber ML, Whitney J, Maile MS, et al. Paternally inherited cis-regulatory structural variants are associated with autism. Science. 2018;360:327–31.
Janecka M, Mill J, Basson MA, Goriely A, Spiers H, Reichenberg A, et al. Advanced paternal age effects in neurodevelopmental disorders—review of potential underlying mechanisms. Translational Psychiatry. 2017;7:e1019.
Acknowledgements
The authors thank the ASD families that participated and the researchers that contributed indirectly to this study: Ana Cristina de Sanctis Girardi, Susan Walker and Giovanna Pellachia. We also thank Dr. André Fujita for support in statistical analysis. We wish to acknowledge support and resources of MSSNG (https://research.mss.ng), Autism Speaks, and The Centre for Applied Genomics, at The Hospital for Sick Children, Toronto, Canada. We also appreciate the contributions of Wilson WL Sung and Darwin D’Souza.
Funding
This work was supported by grants from the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, CEGH-CEL/CEPID process 2013/08028-1, Ph.D. scholarship process 2017/05824-2 and 2018/13743-5), and by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, process 466651/2014-7).
Author information
Authors and Affiliations
Contributions
EMSM and MRPB designed the study. EMSM, GSC, and CISC performed data collection, analysis and helped write the manuscript. SWS conceived and designed MSSNG project and revised the manuscript. FM performed statistical analysis. MS helped with DeNovoGear pipeline and analysis for de novo variants identification. JYTW helped with data annotation and inherited risk variants analysis. AJSC, SLP, WE, BT, and MZ helped perform different components of analysis and data interpretations. MZ revised the manuscript. ECZ contributed to clinical evaluation. NCVL helped with data collection and additional genetic tests of the probands. All authors contributed to data discussion and interpretation of the results.
Corresponding author
Ethics declarations
Competing interests
SWS is on the Scientific Advisory Committee of Population Bio, and serves as a Highly Cited Academic Advisor for the King Abdulaziz University. The remaining authors declare no conflicts of interest.
Ethics approval and consent to participate
The ethics committee of the Instituto de Biociências at Universidade de São Paulo approved the project (accession number 1.133.486), and all the research participants signed a consent term, with written informed consent for publication of individual details obtained.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Costa, C.I.S., da Silva Campos, G., da Silva Montenegro, E.M. et al. Three generation families: Analysis of de novo variants in autism. Eur J Hum Genet 31, 1017–1022 (2023). https://doi.org/10.1038/s41431-023-01398-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-023-01398-6
This article is cited by
-
Characterization of zinc finger protein 536, a neuroendocrine regulator, using pan-cancer analysis
European Journal of Medical Research (2024)
-
Meta-analysis of 46,000 germline de novo mutations linked to human inherited disease
Human Genomics (2024)
-
Genomic analysis of 116 autism families strengthens known risk genes and highlights promising candidates
npj Genomic Medicine (2024)