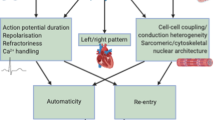
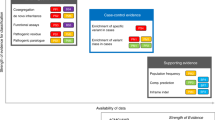
Abstract
A substantial proportion of atrial fibrillation (AF) cases cannot be explained by acquired AF risk factors. Limited guidelines exist that support routine genetic testing. We aim to determine the prevalence of likely pathogenic and pathogenic variants from AF genes with robust evidence in a well phenotyped early-onset AF population. We performed whole exome sequencing on 200 early-onset AF patients. Variants from exome sequencing in affected individuals were filtered in a multi-step process, prior to undergoing clinical classification using current ACMG/AMP guidelines. 200 AF individuals were recruited from St. Paul’s Hospital and London Health Sciences Centre who were ≤ 60 years of age and without any acquired AF risk factors at the time of AF diagnosis. 94 of these AF individuals had very early-onset AF ( ≤ 45). Mean age of AF onset was 43.6 ± 9.4 years, 167 (83.5%) were male and 58 (29.0%) had a confirmed family history. There was a 3.0% diagnostic yield for identifying a likely pathogenic or pathogenic variant across AF genes with robust gene-to-disease association evidence. This study demonstrates the current diagnostic yield for identifying a monogenic cause for AF in a well-phenotyped early-onset AF cohort. Our findings suggest a potential clinical utility for offering different screening and treatment regimens in AF patients with an underlying monogenic defect. However, further work is needed to dissect the additional monogenic and polygenic determinants for patients without a genetic explanation for their AF despite the presence of specific genetic indicators such as young age of onset and/or positive family history.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Data will be made available upon request.
References
Chugh S, Havmoeller R, Narayanan K, Singh D, Rienstra M, Benjamin E, et al. Worldwide epidemiology of atrial fibrillation: A global burden of disease 2010 study. Circulation 2014;129:837–47.
Darbar D. Genetics of atrial fibrillation: Rare mutations, common polymorphisms, and clinical relevance. Heart Rhythm. 2008;5:483–6.
Weng L, Preis S, Hulme O, Larson M, Choi S, Wang B, et al. Genetic predisposition, clinical risk factor burden, and lifetime risk of atrial fibrillation. Circulation 2018;137:1027–38.
Lévy S, Maarek M, Coumel P, Guize L, Lekieffre J, Medvedowsky J, et al. Characterization of different subsets of atrial fibrillation in general practice in France: The ALFA study. The college of French cardiologists. Circulation 1999;99:3028–35.
Kalstø S, Siland J, Rienstra M, Christophersen I. Atrial fibrillation genetics update: Toward clinical implementation. Front Cardiovasc Med. 2019;6:127.
Olesen M, Nielsen M, Haunsø S, Svendsen J. Atrial fibrillation: The role of common and rare genetic variants. Eur J Hum Genet. 2014;22:297–306.
Nattel S, Dobrev D. Controversies about atrial fibrillation mechanisms. Circ Res. 2017;120:1396–8.
Andrade J, Verma A, Mitchell L, Parkash R, Leblanc K, Atzema C, et al. 2018 focused update of the canadian cardiovascular society guidelines for the management of atrial fibrillation. Can J Cardiol 2018;34:1371–92.
Andrade J, Aguilar M, Atzema C, Bell A, Cairns J, Cheung C, et al. The 2020 Canadian cardiovascular society/canadian heart rhythm society comprehensive guidelines for the management of atrial fibrillation. Can J Cardiol. 2020;36:1847–948.
Strande N, Riggs E, Buchanan A, Ceyhan-Birsoy O, Distefano M, Dwight S, et al. Evaluating the clinical validity of gene-disease associations: An evidence-based framework developed by the clinical genome resource. Am J Hum Genet. 2017;100:895–906.
Ingles J, Goldstein J, Thaxton C, Caleshu C, Corty EW, Crowley SB, et al. Evaluating the clinical validity of hypertrophic cardiomyopathy genes. Circulation: Genomic and Precision. Medicine 2019;12:e002460.
Jordan E, Peterson L, Ai T, Asatryan B, Bronicki L, Brown E, et al. Evidence-based assessment of genes in dilated cardiomyopathy. Circulation 2021;144:7–19.
Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: A Fast and Accurate Lllumina paired-end ReAd MergeR. Bioinformatics 2014;30:614–20.
Bolger A, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Lllumina sequence data. Bioinformatics 2014;30:2114–20.
Li H, Durbin R. Fast and accurate long-read alignment with burrows–wheeler transform. Bioinformatics 2010;26:589–95.
Kosugi S, Natsume S, Yoshida K, Maclean D, Cano L, Kamoun S, et al. Coval: Improving alignment quality and variant calling accuracy for next-generation sequencing data. PLoS ONE. 2013;8:e75402.
Cornish A, Guda C. A comparison of variant calling pipelines using genome in a bottle as a reference. Biomed Res Int. 2015;2015:456479.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet Med. 2015;17:405–23.
Chen Y, Xu S, Bendahhou S, Wang X, Wang Y, Xu W, et al. KCNQ1 Gain-of-function mutation in familial atrial fibrillation. Science 2003;299:251–4.
Steffensen AB, Refsgaard L, Andersen MN, Vallet C, Mujezinovic A, Haunsø S, et al. IKs gain- and loss-of-function in early-onset lone atrial fibrillation. J Cardiovasc Electrophysiol. 2015;26:715–23.
Ellinor PT, Nam EG, Shea MA, Milan DJ, Ruskin JN, CA M. Cardiac sodium channel mutation in atrial fibrillation. Heart Rhythm. 2008;5:99–105.
Olson TM, Alekseev AE, Liu XK, Park S, Zingman LV, Bienengraeber M, et al. Kv1.5 Channelopathy Due to KCNA5 loss-of-function mutation causes human atrial fibrillation. Hum Mol Genet. 2006;15:2185–91.
Sébillon P, Bouchier C, Bidot LD, Bonne G, Ahamed K, Charron P, et al. Expanding the phenotype of LMNA mutations in dilated cardiomyopathy and functional consequences of these mutations. J Med Genet. 2003;40:560–7.
Yang YQ, Xu YJ, Li RG, Qu XK, Fang WY, Liu X. Prevalence and Spectrum of PITX2c mutations associated with familial atrial fibrillation. Int J Cardiol. 2013;168:2873–6.
Postma AV, van de Meerakker JB, Mathijssen IB, Barnett P, Christoffels VM, Ilgun A, et al. A Gain-of-Function TBX5 mutation is associated with atypical Holt–Oram syndrome and paroxysmal atrial fibrillation. Circ Res. 2008;102:1433–42.
Gollob MH, Jones DL, Krahn AD, Danis L, Gong XQ, Shao Q, et al. Somatic Mutations in the Connexin 40 Gene (GJA5) in Atrial Fibrillation. N. Engl J Med. 2006;354:2677–88.
Ahlberg GRL, Lundegaard PR, Andreasen L, Ranthe MF, Linscheid N, Nielsen JB, et al. Rare truncating variants in the sarcomeric protein titin associate with familial and early-onset atrial fibrillation. Nat Commun. 2018;9:4316.
Choi SH, Weng LC, Roselli C, Lin H, Haggerty CM, Shoemaker MB, et al. Association between titin loss-of-function variants and early-onset atrial fibrillation. JAMA 2018;320:2354–64.
Goodyer W, Dunn K, Caleshu C, Jackson M, Wylie J, Moscarello T, et al. Broad genetic testing in a clinical setting uncovers a high prevalence of titin loss-of-function variants in very early onset atrial fibrillation. Circ Genom Precis Med. 2019;12:e002713.
Lazarte J, Laksman Z, Wang J, Robinson J, Dron J, Leach E, et al. Enrichment of loss-of-function and copy number variants in ventricular. Cardiomyopathy Genes in “Lone” Atrial Fibrillation. Europace. 2021;23:844–50.
Palmio J, Leonard-Louis S, Sacconi S, Savarese M, Penttila S, Semmler AL, et al. Expanding the importance of HMERF Titinopathy: New mutations and clinical aspects. J Neurol. 2019;266:680–90.
Yoneda Z, Anderson K, Quintana J, O’Neill M, Sims R, Glazer A, et al. Early-onset atrial fibrillation and the prevalence of rare variants in cardiomyopathy and arrhythmia genes. JAMA Cardiol. 2021;6:e213370.
Kelly MA, Caleshu C, Morales A, Buchan J, Wolf Z, Harrison SM, et al. Adaptation and validation of the ACMG/AMP variant classification framework for MYH7-associated inherited cardiomyopathies: Recommendations by ClinGen’s inherited cardiomyopathy expert panel. Genet Med. 2018;20:351–9.
Chalazan B, Mol D, Darbar F, Ornelas-Loredo A, Al-Azzam B, Chen Y, et al. Association of rare genetic variants and early-onset atrial fibrillation in ethnic minority individuals. JAMA Cardiol. 2021;6:811–9.
Ackerman MJ, Priori SG, Willems S, Berul C, Brugada R, Calkins H, et al. HRS/EHRA Expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies. Europace 2011;13:1077–109.
Priori SG, Wilde AA, Horie M, Cho Y, Behr ER, Berul C, et al. HRS/EHRA/APHRS Expert consensus statement on the diagnosis and management of patients with inherited primary arrhythmia syndromes. Heart Rhythm. 2013;10:1932–63.
Shoemaker MB, Shah RL, Roden DM, Perez MV. How will genetics inform the clinical care of atrial fibrillation? Circ Res. 2020;127:111–27.
Funding
This work was supported by the Canadian Cardiovascular Society, University of British Columbia Cardiology Academic Practice Plan; Charles Kerr Scholarship in Cardiovascular Genetics.
Author information
Authors and Affiliations
Contributions
Conceptualization: BC, AL, ZL; Data curation: BC, AL; Formal analysis: BC, AM; Investigation: BC; Resources: AL, ZL; Software: AL; Visualization: BC; Writing-original draft: BC; Writing-review & editing: BC, EL, AL, KR, MB, JW, LH, TR, JL, RH, AL, ZL.
Corresponding author
Ethics declarations
Ethical approval
We attest that the research included in this report was conducted in a manner consistent with the principles of research ethics, such as those described in the Declaration of Helsinki and/or the Belmont Report. In particular, this research was conducted with the voluntary, informed consent of any research participants, free of coercion or coercive circumstances, and received Research Ethics Board (REB) approval from the University of British Columbia that is consistent with the principles of research ethics and the legal requirements of the lead authors’ jurisdiction(s). Written informed consent was obtained from all participants under a protocol approved by the University of British Columbia Research Ethics Board (H16-02531).
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chalazan, B., Freeth, E., Mohajeri, A. et al. Genetic testing in monogenic early-onset atrial fibrillation. Eur J Hum Genet 31, 769–775 (2023). https://doi.org/10.1038/s41431-023-01383-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-023-01383-z
This article is cited by
-
Unusual genomic variants require unusual analyses
European Journal of Human Genetics (2023)