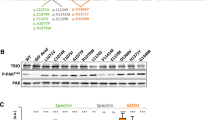
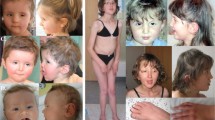
Abstract
RAC1 is a member of the Rac/Rho GTPase subfamily within the RAS superfamily of small GTP-binding proteins, comprising 3 paralogs playing a critical role in actin cytoskeleton remodeling, cell migration, proliferation and differentiation. De novo missense variants in RAC1 are associated with a rare neurodevelopmental disorder (MRD48) characterized by DD/ID and brain abnormalities coupled with a wide range of additional features. Structural and functional studies have documented either a dominant negative or constitutively active behavior for a subset of mutations. Here, we describe two individuals with previously unreported de novo missense RAC1 variants. We functionally demonstrate their pathogenicity proving a gain-of-function (GoF) effect for both. By reviewing the clinical features of these two individuals and the previously published MRD48 subjects, we further delineate the clinical profile of the disorder, confirming its phenotypic variability. Moreover, we compare the main features of MRD48 with the neurodevelopmental disease caused by GoF variants in the paralog RAC3, highlighting similarities and differences. Finally, we review all previously reported variants in RAC proteins and in the closely related CDC42, providing an updated overview of the spectrum and hotspots of pathogenic variants affecting these functionally related GTPases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The exome sequencing data that support the findings of this work are available on request from the corresponding author (MT). The data are not publicly available due to due to privacy/ethical restrictions. The pathogenic variants identified in this work have been submitted to ClinVar (c.475G>A, SCV002605544; c.184G>A, SCV002605545).
References
Hall A. Rho family GTPases. Biochem Soc Trans. 2012;40:1378–82. https://doi.org/10.1042/BST20120103.
Aspenström P, Fransson A, Saras J. Rho GTPases have diverse effects on the organization of the actin filament system. Biochem J. 2004;377:327–37. https://doi.org/10.1042/BJ20031041.
Scala M, Nishikawa M, Ito H, Tabata H, Khan T, Accogli A, et al. Variant-specific changes in RAC3 function disrupt corticogenesis in neurodevelopmental phenotypes. Brain. 2022;145:3308–27. https://doi.org/10.1093/brain/awac106.
Porter AP, Papaioannou A, Malliri A. Deregulation of Rho GTPases in cancer. Small GTPases. 2016;7:123–38. https://doi.org/10.1080/21541248.2016.1173767.
Stankiewicz TR, Linseman DA. Rho family GTPases: key players in neuronal development, neuronal survival, and neurodegeneration. Front Cell Neurosci. 2014;8:314. https://doi.org/10.3389/fncel.2014.00314.
Scala M, Nishikawa M, Nagata KI, Striano P. Pathophysiological mechanisms in neurodevelopmental disorders caused by RacGTPases dysregulation: what’s behind Neuro-RACopathies. Cells. 2021;10:3395. https://doi.org/10.3390/cells10123395.
Reijnders MRF, Ansor NM, Kousi M, Yue WW, Tan PL, Clarkson K, et al. RAC1 missense mutations in developmental disorders with diverse phenotypes. Am J Hum Genet. 2017;101:466–77. https://doi.org/10.1016/j.ajhg.2017.08.007.
Barbosa S, Greville-Heygate S, Bonnet M, Godwin A, Fagotto-Kaufmann C, Kajava AV, et al. Opposite modulation of RAC1 by mutations in TRIO is associated with distinct, domain-specific neurodevelopmental disorders. Am J Hum Genet. 2020;106:338–55. https://doi.org/10.1016/j.ajhg.2020.01.018.
Moll J, Sansig G, Fattori E, van der Putten H. The murine rac1 gene: cDNA cloning, tissue distribution and regulated expression of rac1 mRNA by disassembly of actin microfilaments. Oncogene. 1991;6:863–6. PMID: 1905006.
Shirsat NV, Pignolo RJ, Kreider BL, Rovera G. A member of the ras gene superfamily is expressed specifically in T, B and myeloid hemopoietic cells. Oncogene. 1990;5:769–72. PMID: 2189110.
Malosio ML, Gilardelli D, Paris S, Albertinazzi C, de Curtis I. Differential expression of distinct members of Rho family GTP-binding proteins during neuronal development: identification of Rac1B, a new neural-specific member of the family. J Neurosci. 1997;17:6717–28. https://doi.org/10.1523/JNEUROSCI.17-17-06717.1997.
Costain G, Callewaert B, Gabriel H, Tan TY, Walker S, Christodoulou J, et al. De novo missense variants in RAC3 cause a novel neurodevelopmental syndrome. Genet Med. 2019;21:1021–6. https://doi.org/10.1038/s41436-018-0323-y.
Lougaris V, Baronio M, Gazzurelli L, Benvenuto A, Plebani A. RAC2 and primary human immune deficiencies. J Leukoc Biol. 2020;108:687–96. https://doi.org/10.1002/JLB.5MR0520-194RR.
Lelieveld SH, Reijnders MR, Pfundt R, Yntema HG, Kamsteeg EJ, de Vries P, et al. Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat Neurosci. 2016;19:1194–6. https://doi.org/10.1038/nn.4352.
Haugh IM, Pineider JL, Agim NG. Ichthyosiform changes in a patient with RAC1 mutation. Pediatr Dermatol. 2021;38:1590–1. https://doi.org/10.1111/pde.14844.
Tian C, Kay Y, Sadybekov A, Rao S, Katritch V, Herring BE. An intellectual disability-related missense mutation in Rac1 prevents LTP induction. Front Mol Neurosci. 2018;11:223 https://doi.org/10.3389/fnmol.2018.00223.
Martinelli S, Krumbach OHF, Pantaleoni F, Coppola S, Amin E, Pannone L, et al. Functional dysregulation of CDC42 causes diverse developmental phenotypes. Am J Hum Genet. 2018;102:309–20. https://doi.org/10.1016/j.ajhg.2017.12.015.
Lam MT, Coppola S, Krumbach OHF, Prencipe G, Insalaco A, Cifaldi C, et al. A novel disorder involving dyshematopoiesis, inflammation, and HLH due to aberrant CDC42 function. J Exp Med. 2019;216:2778–99. https://doi.org/10.1084/jem.20190147.
Coppola S, Insalaco A, Zara E, Di Rocco M, Marafon DP, Spadaro F, et al. Mutations at the C-terminus of CDC42 cause distinct hematopoietic and autoinflammatory disorders. J Allergy Clin Immunol. 2022;150:223–8. https://doi.org/10.1016/j.jaci.2022.01.024.
Daimon E, Shibukawa Y, Thanasegaran S, Yamazaki N, Okamoto N. Macrothrombocytopenia of Takenouchi-Kosaki syndrome is ameliorated by CDC42 specific- and lipidation inhibitors in MEG-01 cells. Sci Rep. 2021;11:17990. https://doi.org/10.1038/s41598-021-97478-y.
Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–5. https://doi.org/10.1038/nmeth.2089.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. https://doi.org/10.1038/gim.2015.30.
Frazer J, Notin P, Dias M, Gomez A, Min JK, Brock K, et al. Disease variant prediction with deep generative models of evolutionary data. Nature. 2021;599:91–95. https://doi.org/10.1038/s41586-021-04043-8.
Chang MT, Asthana S, Gao SP, Lee BH, Chapman JS, Kandoth C, et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol. 2016;34:155–63. https://doi.org/10.1038/nbt.3391.
Majole´e J, Podieh F, Hordijk PL, Kovačević I. The interplay of Rac1 activity, ubiquitination and GDI binding and its consequences for endothelial cell spreading. PLoS One. 2021;16:e0254386. https://doi.org/10.1371/journal.pone.0254386.
Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–10. https://doi.org/10.1016/0092-8674(92)90164-8.
Carta C, Pantaleoni F, Bocchinfuso G, Stella L, Vasta I, Sarkozy A, et al. Germline missense mutations affecting KRAS Isoform B are associated with a severe Noonan syndrome phenotype. Am J Hum Genet. 2006;79:129–35. https://doi.org/10.1086/504394.
Janakiraman M, Vakiani E, Zeng Z, Pratilas CA, Taylor BS, Chitale D, et al. Genomic and biological characterization of exon 4 KRAS mutations in human cancer. Cancer Res. 2010;70:5901–11. https://doi.org/10.1158/0008-5472.CAN-10-0192.
Gremer L, Merbitz-Zahradnik T, Dvorsky R, Cirstea IC, Kratz CP, Zenker M, et al. Germline KRAS mutations cause aberrant biochemical and physical properties leading to developmental disorders. Hum Mutat. 2011;32:33–43. https://doi.org/10.1002/humu.21377.
Sanghavi HM, Rashmi R, Dasgupta A, Majumdar S. G-domain prediction across the diversity of G protein families. bioRxiv. 2019;24:888222. https://doi.org/10.1101/2019.12.24.888222.
Hiraide T, KabaYasui H, Kato M, Nakashima M, Saitsu H. A de novo variant in RAC3 causes severe global developmental delay and a middle interhemispheric variant of holoprosencephaly. J Hum Genet. 2019;64:1127–32. https://doi.org/10.1038/s10038-019-0656-7.
Sugihara K, Nakatsuji N, Nakamura K, Nakao K, Hashimoto R, Otani H, et al. Rac1 is required for the formation of three germ layers during gastrulation. Oncogene. 1998;17:3427–33. https://doi.org/10.1038/sj.onc.1202595.
He F, Soriano P. A critical role for PDGFRα signaling in medial nasal process development. PLoS Genet. 2013;9:e1003851. https://doi.org/10.1371/journal.pgen.1003851.
Chai Y, Jiang X, Ito Y, Bringas P Jr, Han J, Rowitch DH, et al. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–9. https://doi.org/10.1242/dev.127.8.1671.
Leung C, Engineer A, Kim MY, Lu X, Feng Q. Myocardium-specific deletion of Rac1 causes ventricular noncompaction and outflow tract defects. J Cardiovasc Dev Dis. 2021;8:29. https://doi.org/10.3390/jcdd8030029.
Allanson JE, Cunniff C, Hoyme HE, McGaughran J, Muenke M, Neri G. Elements of morphology: standard terminology for the head and face. Am J Med Genet A. 2009;149A:6–28. https://doi.org/10.1002/ajmg.a.32612.
Pennucci R, Talpo F, Astro V, Montinaro V, Morè L, Cursi M, et al. Loss of either Rac1 or Rac3 GTPase differentially affects the behavior of mutant mice and the development of functional GABAergic networks. Cereb Cortex. 2016;26:873–90. https://doi.org/10.1093/cercor/bhv274.
Mosaddeghzadeh N, Ahmadian MR. The RHO family GTPases: mechanisms of regulation and signaling. Cells. 2021;10:1831. https://doi.org/10.3390/cells10071831.
Fiegen D, Haeusler LC, Blumenstein L, Herbrand U, Dvorsky R, Vetter IR, et al. Alternative splicing of Rac1 generates Rac1b, a self-activating GTPase. J Biol Chem. 2004;279:4743–9. https://doi.org/10.1074/jbc.M310281200.
Asiri A, Alwadaani D, Umair M, Alhamoudi KM, Almuhanna MH, Nasir A, et al. Pancytopenia, recurrent infection, poor wound healing, heterotopia of the brain probably associated with a candidate novel de novo CDC42 gene defect: expanding the molecular and phenotypic spectrum. Genes 2021;12:294. https://doi.org/10.3390/genes12020294.
Tartaglia M, Gelb BD. Disorders of dysregulated signal traffic through the RAS-MAPK pathway: phenotypic spectrum and molecular mechanisms. Ann N Y Acad Sci. 2010;1214:99–121. https://doi.org/10.1111/j.1749-6632.2010.05790.x.
Niihori T, Aoki Y, Okamoto N, Kurosawa K, Ohashi H, Mizuno S, et al. HRAS mutants identified in Costello syndrome patients can induce cellular senescence: possible implications for the pathogenesis of Costello syndrome. J Hum Genet. 2011;56:707–15. https://doi.org/10.1038/jhg.2011.85.
Zampino G, Pantaleoni F, Carta C, Cobellis G, Vasta I, Neri C, et al. Diversity, parental germline origin, and phenotypic spectrum of de novo HRAS missense changes in Costello syndrome. Hum Mutat. 2007;28:265–72. https://doi.org/10.1002/humu.20431.
Gripp KW, Lin AE. Costello syndrome: a Ras/mitogen activated protein kinase pathway syndrome (rasopathy) resulting from HRAS germline mutations. Genet Med. 2012;14:285–92. https://doi.org/10.1038/gim.0b013e31822dd91f.
Harms FL, Kloth K, Bley A, Denecke J, Santer R, Lessel D, et al. Activating mutations in PAK1, encoding p21-activated kinase 1, cause a neurodevelopmental disorder. Am J Hum Genet. 2018;103:579–91. https://doi.org/10.1016/j.ajhg.2018.09.005.
Hori K, Shimaoka K, Hoshino M. AUTS2 gene: keys to understanding the pathogenesis of neurodevelopmental disorders. Cells. 2021;11:11. https://doi.org/10.3390/cells11010011.
Duffney LJ, Wei J, Cheng J, Liu W, Smith KR, Kittler JT, et al. Shank3 deficiency induces NMDA receptor hypofunction via an actin-dependent mechanism. J Neurosci. 2013;33:15767–78. https://doi.org/10.1523/JNEUROSCI.1175-13.2013.
Dong T, He J, Wang S, Wang L, Cheng Y, Zhong Y. Inability to activate Rac1-dependent forgetting contributes to behavioral inflexibility in mutants of multiple autism-risk genes. Proc Natl Acad Sci USA. 2016;113:7644–9. https://doi.org/10.1073/pnas.1602152113.
Jones KL, Jones MC, and Del Campo M. Smith’s recognizable patterns of human malformation. Philadelphia, PA: Elsevier Health Sciences; 2013.
Acknowledgements
We are grateful to the families who participated in this study.
Funding
This work was supported by the Italian Ministry of Health (5×1000_2019 and CCR-2017-23669081 to MT, and RF-2018-12366931 and GR-2019-12371203 to FCR), Istituto Superiore di Sanità (Bando Ricerca Indipendente ISS 2020-2022-ISS20-39c812dd2b3c to SC), Fondazione AIRC per la Ricerca sul Cancro (IG 21614, to MT), and Italian Ministry of Research (FOE_2019 to MT).
Author information
Authors and Affiliations
Contributions
MP and MT conceived the work, and wrote the manuscript. AC, CM, FP, VC, LC and MN performed the genomic analyses and analyzed and validated the genomic data. EB performed the structural analyses. MP, FCR, EA, CM and DM collected the clinical data and contributed to the clinical data analyses. EZ, FS, and SC designed and performed the functional analyses. All coauthors helped write and revise the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The study was approved by the local Institutional Ethical Committee (ref. 1702_OPBG_2018). Clinical data, pictures, DNA samples and other biological specimens were collected, used, and stored after signed informed consents from the participating subjects/families were secured. Permission to publish the clinical pictures and the clinical details was obtained for both individuals.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Priolo, M., Zara, E., Radio, F.C. et al. Clinical profiling of MRD48 and functional characterization of two novel pathogenic RAC1 variants. Eur J Hum Genet 31, 805–814 (2023). https://doi.org/10.1038/s41431-023-01351-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-023-01351-7
This article is cited by
-
Structural rearrangements as a recurrent pathogenic mechanism for SETBP1 haploinsufficiency
Human Genomics (2024)
-
Unusual genomic variants require unusual analyses
European Journal of Human Genetics (2023)