Abstract
Despite substantial efforts in identifying both rare and common variants affecting disease risk, in the majority of diseases, a large proportion of unexplained genetic risk remains. We propose that variable number tandem repeats (VNTRs) may explain a proportion of the missing genetic risk. Herein, in a pilot study with a retrospective cohort design, we tested whether VNTRs are causal modifiers of breast cancer risk in 347 female carriers of the BRCA1 185delAG pathogenic variant, an important group given their high risk of developing breast cancer. We performed targeted-capture to sequence VNTRs, called genotypes with adVNTR, tested the association of VNTRs and breast cancer risk using Cox regression models, and estimated the effect size using a retrospective likelihood approach. Of 303 VNTRs that passed quality control checks, 4 VNTRs were significantly associated with risk to develop breast cancer at false discovery rate [FDR] < 0.05 and an additional 4 VNTRs had FDR < 0.25. After determining the specific risk alleles, there was a significantly earlier age at diagnosis of breast cancer in carriers of the risk alleles compared to those without the risk alleles for seven of eight VNTRs. One example is a VNTR in exon 2 of LINC01973 with a per-allele hazard ratio of 1.58 (1.07–2.33) and 5.28 (2.79–9.99) for the homozygous risk-allele genotype. Results from this first systematic study of VNTRs demonstrate that VNTRs may explain a proportion of the unexplained genetic risk for breast cancer.
Similar content being viewed by others
Introduction
For carriers of pathogenic variants (PVs) in BRCA1, the lifetime risk for developing breast cancer (up to 80% lifetime risk) is a six-fold increase over that of average risk women and ovarian cancer risk (up to a 44% lifetime risk) is up to a 30-fold increase [1]. Despite these substantially elevated risks, penetrance is incomplete (not all carriers will develop cancer) and age at cancer diagnosis varies. The limited understanding of factors that modify cancer risks in BRCA1 carriers hampers clinical decision-making ability, including decisions about the appropriate type and timing of risk reducing surgeries. Therefore, there is a critical, clinically relevant need for more refined risk estimates.
The variation in risk, even in identical PVs carriers, suggests that modifier factors, both genetic and environmental, affect cancer risks [2]. Studies to identify “modifier genes” that govern the phenotypic expression of BRCA PV carriers have been ongoing since the early 2000’s, conducted largely through the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA) [3, 4]. Through genome-wide association studies (GWAS), single nucleotide polymorphisms (SNPs) have been identified that, when combined into a polygenic risk score (PRS), better define BRCA1 carriers at higher and lower risk of developing breast cancer (e.g. [5,6,7,8]). However, these modifier variants are estimated to explain only ~8% of the familial risk in BRCA1 carriers [8,9,10,11]. Identifying additional genetic modifiers will facilitate better risk estimates for clinical decision-making on timing and options for risk reduction.
Variable number tandem repeats (VNTRs) may plausibly account for some of the missing genetic risk. They are known to modulate biologic processes, including gene expression and protein function [12,13,14,15,16]. These eVNTRs (VNTR expression Quantitative Trait Loci) also mediate risks of developing various cancers [17, 18] including breast cancer [19,20,21,22]. A genome-wide investigation of VNTRs as modifiers has been hampered by technical difficulties; however, adVNTR [12, 23] became available to genotype VNTRs (i.e., count repeat units) from next generation sequencing (NGS) data. This tool uses Hidden Markov models (HMM) to model each VNTR, count repeat units, and detect sequence variation.
In this pilot study using a retrospective cohort design, we tested a new paradigm – that VNTRs are causal modifiers of breast cancer risk. They have not been systematically investigated as they are poorly tagged by nearby SNPs [14]. Previous GWAS conducted through CIMBA have demonstrated heterogeneity of breast cancer risk by type of variant and variant location in BRCA1/2, breast tumor subtypes, and race and ethnicity [6, 10, 24,25,26,27]. Therefore, to reduce potential confounding with unmeasured variables, we tested the association in carriers of a single recurring PV in BRCA1. We performed targeted-capture to sequence VNTRs, called genotypes with adVNTR, and explored the association of VNTRs and breast cancer in 327 women carrying the pathogenic BRCA1 185delAG mutation [NM_007294.4(BRCA1):c.68_69del (p.Glu23fs) (rs80357914)].
Methods
Participants
Females over the age of 18 years of age carrying the pathogenic BRCA1 variant 185delAG (NM_007294.3:c.66_67del) were eligible. Of the 347 participants with DNA, 250 were enrolled at the Sheba Medical Center (SMC) in Israel. All participants underwent oncogenetic counseling and genotyping of cancer susceptibility genes, including BRCA1. Referral to the oncogenetics services came from several sources: women who developed breast and/or ovarian cancer (consecutive women at the SMC) (n = 57), cancer-free women with a significant family history of breast and /or ovarian cancer (n = 61) or a known mutation in their family (n = 125), and from population screens of the three predominant mutations in Ashkenazi Jewish (AJ) women in BRCA1 and BRCA2 (n = 7) [28], a procedure recently approved and included in the Israeli “health basket” for all AJ women as a screening procedure with no need for pre-test counseling. Another 95 participants were enrolled into the Clinical Cancer Genomics Community Research Network (CCGCRN) housed at the City of Hope in which eligibility was any individual receiving genetic cancer risk assessment (GCRA) and specific for this study, a diagnosis of invasive breast cancer. Another two participants were recruited and enrolled in a research study of women in high-risk breast cancer families. Only the proband was selected from a pedigree so that none of the participants were related. All participants provided written informed consent under IRB-approved protocols at their respective institutions. There was no follow-up of participants nor data available for additional risk factors. None of the participants had prophylactic surgeries.
VNTR genotyping
VNTR selection
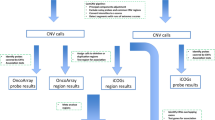
To get an initial list of VNTRs (of four or more base pair repeats), Tandem Repeat Finder (TRF) [29] was applied to the human reference genome [GRCh38], and 559,804 VNTRs were identified. To focus on the most relevant candidates, we selected VNTRs that intersected with coding exons, promoters, or untranslated regions (UTRs) of genes in RefSeq (https://www.ncbi.nlm.nih.gov/refseq/). VNTRs were excluded if they were located in low-complexity sequence (e.g. close to a telomere) resulting in 8953 candidate VNTRs. Lastly, only candidate VNTRs with total length of 140 bp or shorter (n = 6271) were included so that genotypes could be confidently assigned with Illumina short read sequencing data. We used the Agilent SureDesign software to design probes for 6271 VNTRs. Of these 6271 VNTRs, 1398 are in coding exons, 2000 are in promoter regions, and 2873 are in UTRs. We observed that 85 VNTRs were in repetitive DNA regions where no probes could be designed and 21 were on the Y chromosome. Excluding these 106 VNTRs and using the least stringent parameters, probes were designed to cover 6165 VNTRs.
Library preparation and targeted-capture DNA sequencing and processing of reads
Details are provided in Supplementary Methods. Briefly, Illumina sequencing libraries were created from 500 ng DNA using KAPA Hyper (KAPA Biosystems) reagents along with our optimized protocols [30, 31]. Sequence reads were aligned to NCBI build GRCh38 using Burrows-Wheeler Aligner (BWA). From the BAM files, genotypes from VNTRs were assigned using adVNTR-NN adapted from adVNTR [23] based on minimal total supporting reads ≥10 and minimal proportion of reads to support alternative allele ≥ 0.25.
Confirmation of VNTR genotyping results from adVNTR
Using the unique flanking regions of the selected VNTRs, PCR primers were designed to amplify 50 ng DNA from up to 4 samples per VNTR genotype. PCR reactions were performed using Taq polymerase (Qiagen) and amplification was confirmed using gel electrophoresis. Samples were then sequenced on an Applied Biosystems SeqStudio Genetic Analyzer (ThermoFisher Scientific).
VNTR sequences were visualized using Quality Check and Variant Analysis Modules on the ThermoFisher Cloud. The visualized sequence in conjunction with the product sizes from the post-PCR gel electrophoresis were used to verify genotyping calling made by adVNTR. For homozygotes, this was done by observing a single band of the correct size during gel electrophoresis and by quality sequence for the number of repeats called by adVNTR. Whereas heterozygotes were confirmed by observing multiple bands of expected size differentials on the gel and a poor-quality Sanger sequence at the point of allele differences.
Statistical analysis
After genotypes were assigned for each VNTR, we tested for Hardy-Weinberg equilibrium (HWE) [32]. For those that were in HWE (p > 0.001), we tested the association of the VNTR and risk to develop breast cancer using Cox regression models. For each VNTR associated with risk to develop breast cancer, we determined the risk allele group and estimated the hazard ratio using the retrospective likelihood approach (described below). In these analyses, women with a first breast cancer were considered as affected with time to breast cancer diagnosis as the end point; those unaffected with any cancer were censored at age at genetic testing (which also is the date of study entry), and those diagnosed with ovarian cancer prior to breast cancer were censored at age at ovarian cancer diagnosis. There were too few cases of ovarian cancer for analysis.
In the primary analysis, we tested the association between each VNTR marker as a continuous variable and disease risk. Three separate VNTR genotypes were constructed: 1) the average length of the two alleles; 2) the length of only the shorter allele; and 3) the length of only the longer allele [33]. Analyses were adjusted for sample collection site (US or Israel). Probability values were adjusted for multiple comparisons using the False Discovery Rate (FDR) method of Benjamini and Hochberg [34].
For VNTRs with associations at FDR < 0.25 in the primary association analysis, a second analysis was performed to identify the specific risk groups of repeat alleles using a sliding window method [33]. Specifically, for a multi-allele VNTR, a threshold T along the number of repeats from short to long was used to dichotomize allele lengths. An allele was denoted as ‘short’ if it had shorter than T repeat motifs, and ‘long’ otherwise. Multiple values of threshold T were chosen for association tests. For each specific threshold T, the VNTR genotype of an individual was converted to homozygous-short-allele genotype (S/S), heterozygous-short-and-long-allele genotype (S/L), or homozygous-long-allele genotype (L/L). The optimal threshold (cut-point) for each VNTR was determined by choosing T that provided the smallest p-value among the multiple association tests. This cut-point then was used to estimate the hazard ratio using the retrospective likelihood method in order to mitigate potential bias in estimating hazard ratios arising from over-sampling of breast cancer cases [35]. Kaplan–Meier (KM) curves and log-rank tests were used to graphically examine differences in the cumulative probability of breast cancer risk among VNTR genotype groups categorized using the critical cut points for risk alleles. The implementation of Cox regressions and KM analysis was based on relevant functions in R packages of survival and survminer [36]; the retrospective likelihood tests were performed by the “RetroLike_Release_1_0_3” program [35].
Luciferase assays
We conducted luciferase assays to test alleles of one VNTR to determine if it affected expression. We selected the VNTR with the lowest FDR that was in a promoter or 5’UTR region. Details are provided in Supplementary Methods. Briefly, the cloning of VNTR alleles, construction of luciferase reporter plasmids, and measurement of the relative luciferase activities of the plasmid constructs were conducted based on our optimized protocols published previously [37]. All transfections were performed in quadruplicate, and each construct was tested in three independent experiments. The average of 12 relative luciferase measurements for each allele were expressed as the mean ±standard error of mean (SEM). Difference in relative activity values between the risk repeat allele group and reference repeat allele group was tested by one-way ANOVA analysis. The P-value was adjusted for multiple testing using the Tukey’s method [38]; adjusted p- values less than 0.05 were considered as statistically significant.
Results
Participants
The cancer status and ages at diagnosis or enrollment (for non-cancer cases) are shown in Table 1. Of the 347 women, ages ranged from 18 to 77 years with 47.3% having been diagnosed with a first breast cancer, of which 3.5% also were diagnosed with ovarian cancer. The median age at first breast cancer diagnosis was 42 years and the median age of the unaffected group was 47 years.
VNTR genotyping
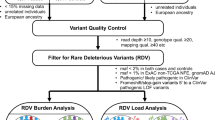
In total, we sequenced 6165 VNTRs in 347 BRCA1 185delAG PV carriers. Genotypes were called using adVNTR-NN. In Fig. 1, the flow diagram of steps for elimination of VNTRs and samples is shown. Of 6165 VNTRs, 3847 (62.4%) VNTRs were removed due to missing more than 5% of genotypes, with the main reasons being VNTRs located in GC-rich regions which had poor amplification during library generation, imperfect repeats, or flanked by other repetitive elements. Another 1622 VNTRs were removed because they were monomorphic (1588 VNTRs) or not in HWE (P value < 0.001; 34 VNTRs). Lastly, 393 VNTRs had heterozygosity <0.02. Because this is a homogeneous dataset of Ashkenazi Jewish ancestry, it was expected that more VNTRs would be monomorphic and within VNTRs, not all alleles would be present. Twenty samples were removed that had more than 10% missing genotypes leaving 327 samples for analysis. The summary of repeat alleles in this dataset for the 303 VNTRs is shown in Table 2.
Association of VNTRs and risk of developing cancer
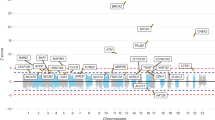
In the primary analysis, we used Cox proportional hazards models to evaluate the association between each VNTR and risk of developing breast cancer, considering the VNTR as a continuous variable. Of 303 VNTRs analyzed, four VNTRs had FDR < 0.05, and an additional four had FDR < 0.25 (Table 3; Supplementary Table 1). The alleles for each of the eight VNTRs were accurately called, with 100% consistency among the adVNTR, agarose gel, and Sanger sequencing results (VNTR 558420 is shown as an example in Supplementary Fig. 1). We then conducted the secondary analysis for the eight VNTRs to identify the specific risk repeat alleles contributing to the significant association. Of the eight VNTRs, six VNTRs had two major repeat alleles (Supplementary Table 1) and therefore only one cutpoint for short or long risk alleles; VNTR 412033 and VNTR 945060 had more than one possible cut-point with the critical cut point determined from the smallest p value in a per-allele trend test (Supplementary Table 2). For seven of eight VNTRs, there was a significant (P < 0.05; per-allele trend test) association of the dichotomized risk allele and breast cancer risk (Table 4). For VNTR 47260, although breast cancer risk increased with repeat length based on the linear trend test (FDR = 0.035), there were too few long repeat alleles (> 9 R) for a stable estimate the hazard ratio. Using CIMBA summary statistics data (https://cimba.ccge.medschl.cam.ac.uk/) for BRCA1 carriers, we examined the association of GWAS SNPs within 200 Kb of each of the 8 VNTRs (n = 9181) in Table 3 (100 Kb left and 100 Kb right of VNTR) and breast cancer risk. None of the SNPs were genome-wide significance (all were p > 10(−5)).
Kaplan–Meier (KM) curves were used to graphically show the difference in the cumulative probability of breast cancer risk for the VNTR genotype groups (Fig. 2 and Supplementary Fig. 2). Individuals with the risk genotypes had significantly earlier ages at diagnosis of breast cancer (log-rank p value < 0.05) (Fig. 2 and Supplementary Fig. 2). For example, the median ages at breast cancer diagnosis for carriers with the S/S genotype and the L/L genotype in VNTR357331 were 40 years and 56 years, respectively (log-rank p value of 0.0014, Fig. 2), indicating the age-modifying effect of breast cancer diagnosis among carriers harboring risk genotype (S/S).
The age at breast cancer diagnosis is on the X-axis and proportion of participants diagnosed with breast cancer is on the Y-axis. The horizontal/vertical dash line is the median age at diagnosis of breast cancer. In this step function of breast cancer risk over age, in panel A, B, and C, the Kaplan–Meier curves for each of the three VNTRs with FDR < 0.05 are shown. Panel A for VNTR253688, 5 and 9 repeats are short (S) alleles; 10 repeats is long (L) and risk allele. Panel B for VNTR357331, 4 repeats is short (S) and risk allele; 5 and 6 repeats are long (L) alleles. Panel C for VNTR412033, 7, 8 and 9 repeats are short (S) and risk alleles; 10 and 11 repeats are long (L) alleles. For each of the VNTRs, there were significantly different risks of developing breast cancer by VNTR genotypes (log-rank p value < 0.05).
Effect of VNTR alleles on expression
For testing the effect on gene expression, we selected the VNTR with the lowest FDR that was located in a gene promoter or 5’ UTR. We tested VNTR 558420 located in the 5’ UTR of ZNF501 (p-value = 0.0025 and FDR = 0.135) (Table 3 and Supplementary Fig. 3) with repeats of 2 R, 3 R and 4 R and 3 genotypes (3 samples with genotype 2/3, 309 with 3/3, and 6 with 3/4). In Fig. 3, normalized luciferase activity is shown for the 2 R, 3 R, 4 R repeats and the control (empty vector) with standard error bars on the top of each group mean. There was a significant (adjusted p value < 0.05) difference between the 2 R and 4 R groups with the 3 R intermediate (Fig. 3) and a significant linear trend of decreased luciferase activity with increasing number of repeats (p = 0.021) (Supplementary Figure 4).
Each experimental group is composed of 12 data points. Data represent fold change in the repeat group relative to vector group, with standard error bar shown for each group. Significance was assessed by one-way ANOVA with pairwise t test and P-value adjusted by the Tukey’s method. Asterisk above standard error bar indicates significance test between the repeat group and vector group; asterisk above the line indicates the significance between the 2 R and 4 R repeat groups; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Discussion
Our study is the first to conduct a systematic study of VNTRs and association with risk to develop cancer in high-risk BRCA1 PV carriers. We identified four VNTRs significantly associated with risk of developing breast cancer in women carrying the 185delAG BRCA1 PV (FDR < 0.05) and another four VNTRs associated with FDR < 0.25.
None of the small number of previous association studies of risk of developing breast cancer and VNTRs at candidate genes had investigated the eight VNTRs we identified. Krontiris and coworkers reported an association of rare alleles in a HRAS1 VNTR and development of cancers, including breast cancer [19], and a meta-analysis of 13 breast cancer studies found an association with breast cancer risk [39]. Functional analysis showed that this HRAS VNTR altered CpG DNA methylation [40]. In a meta-analysis of 17 studies of a CAG-repeat polymorphism in the androgen receptor, they found an association of longer CAG repeats with an increased risk of breast cancer in Caucasian women [41]. In a meta-analysis of two studies of the MNS16A VNTR in the hTERT promoter, they found a significant association with development of breast cancer. In a Japanese study of an 18 bp VNTR in the promoter of PTTG1IP, they found a signficant association with risk of estrogen-receptor positive breast cancer, with functional analysis showing that an increase in the number of repeats increased the binding affinity of ER-alpha [22]. In a study of a VNTR in the promoter of XRCC5, they found a significant association with age at breast cancer diagnosis [20]. None of these VNTRs were included in our analysis. We did not include trinucleotide repeats (AR repeats) in our targeted sequencing and the hRAS and XRCC5 VNTR total lengths were larger than our cut-off size of 140 bp. The MSN1 VNTR was monomorphic and the PTTG1IP VNTR was missing too many genotypes in our set and thus were excluded.
Of the eight VNTRs that we found to be associated with risk of developing breast cancer in this population, several warrant further investigation. VNTR 945060 is in the 5’UTR of ERCC6L, a DNA helicase. ERCC6L is highly expressed in breast tissue and higher levels of expression have been associated with worse survival [42]; silencing of ERCC6L in breast cell lines significantly inhibited cell proliferation [42, 43]. A second VNTR, 253688, is located 3’ of FLJ22447, a lncRNA located near HIF-1α. In a study of esophageal squamous cell carcinoma and gastric cancers to determine the effect of FLJ22447 on HIF-1α, they observed that low expression of lncRNA was associated with expression of HIF-1α suggesting that FLJ22447 may have a regulatory function on HIF-1α expression [44]. High over-expression of HIF-1α is common in breast cancers and is particularly common in BRCA1 carriers [45,46,47]. This VNTR may alter risk to develop breast through affecting HIF-1α.
Given the reports that there are shared genetic contributions between breast cancer and schizophrenia [48], it is interesting that three of the VNTRs are at or in genes (SYN2, ZNF501, ZNF804A) associated with risk to develop schizophrenia [49,50,51,52,53]; VNTR 549198 is in exon 12 of SYN2; VNTR 472060 is in exon 4 of ZNF804A; and VNTR 558420 is in the 5’UTR of ZNF501 and all are most commonly expressed in brain (proteinatlas.org). From our luciferase assays, there was differential expression from varying alleles in the VNTR in the 5’UTR of ZNF501; expression differences for this VNTR were only associated with brain tissue in GTEX [12]. The exonic VNTRs in SYN2 and in ZNF804A cause expansions of poly-serine (Supplementary Fig. 5) and poly-alanine (Supplementary Fig. 6) tracts, respectively. VNTR expansions in gene coding regions have been associated with multiple diseases [54]. Further investigation is needed to assess possible roles in development of breast cancer.
This was a pilot study to determine the feasibility of conducting targeted sequencing of VNTRs and investigating the association of VNTRs as modifiers of disease risk, similar to what has been accomplished with SNPs [11, 24]. We purposefully included women carrying the specific BRCA1 185delAG Ashkenazi Jewish founder PV to try to explain the known variation in risk in women carrying this PV and to reduce potential confounding with unmeasured variables; however, the consequence is that it reduced the number of VNTRs that were polymorphic and restricted the sample size. A second limitation of the study is the small sample size such that estimates of risk are not precise and may be inflated for the rarer risk alleles. In hindsight, using targeted capture and sequencing of 250 bp reads limited the size of repeats and reduced the number of VNTRs that made it through all the quality control checks due to poor amplification of VNTRs in GC-rich regions, difficulty in aligning VNTRs with imperfect repeats and/or with low complexity/repetitive sequence in the flanking regions. However, this pilot study has provided information for future studies. In regards to genotyping VNTRs, longer reads are necessary in order to capture additional VNTRs and a different technology such as whole-genome sequencing (WGS) long-read sequences such as performed by PacBio is needed to overcome issues of sequencing GC-rich regions. With the availability of WGS data in public databases such as the UK Biobank and the All of Us Research Program in the United States, we will be able to assess the association of VNTRs in overall breast cancer and not restricted to this small set. Based on our results herein, we have a better sense of sample size to detect statistically significant associations.
BRCA1 breast cancers are generally basal, triple-negative hormone receptor cancers (TNBC). We have seen from SNP studies of both BRCA1 carriers and women with TNBC that there are fewer SNPs associated with risk than for estrogen-receptor positive breast cancers; SNPs explain approximately 8% of the familial risk in BRCA1 carriers [10]. Thus, identification of VNTRs significantly associated with risk of developing breast cancer in this genetically and ethnically homogeneous population is encouraging; several of which have been observed to play a role in breast cancer. The per-allele HRs for the dichotomized risk alleles in these VNTRs ranged from 1.6 to 6.6 (Table 4) whereas per-allele HRs for SNPs ranged from 1.01 to 1.40 [11, 55], suggesting that VNTRs may have larger effects than SNPs. For the rare VNTR allele in the 5’UTR of ZNF501, we did show that it affected expression. Several reports, including our own, have shown that VNTR motif change have a larger, causal effect on gene expression and function than SNPs [12, 56,57,58]. The relatively large hazard ratios observed in this study need to be validated in larger datasets that include women of diverse ethnicities, a wider spectrum of BRCA1 PVs, and carriers of BRCA2 PVs. Moreover, a larger genome-wide VNTR association study may identify additional VNTRs. In a future study, after identifying and replicating VNTRs associated with risk of developing breast cancer, incorporation into PRS will be warranted.
In summary, the results from this study demonstrate that VNTRs may explain a proportion of the unexplained genetic risk for disease. Similar to SNPs, VNTRs significantly associated with the disease of interest could be incorporated into polygenic risk scores (PRS) to test for improved risk assessment and clinical applicability.
Data availability
Data generated as part of this study are available from the corresponding author on reasonable request.
References
Kuchenbaecker KB, Hopper JL, Barnes DR, Phillips KA, Mooij TM, Roos-Blom MJ, et al. Risks of Breast, Ovarian, and Contralateral Breast Cancer for BRCA1 and BRCA2 Mutation Carriers. JAMA. 2017;317:2402–16.
Levy-Lahad E, Friedman E. Cancer risks among BRCA1 and BRCA2 mutation carriers. Br J Cancer. 2007;96:11–5.
Antoniou AC, Wang X, Fredericksen ZS, McGuffog L, Tarrell R, Sinilnikova OM, et al. A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor-negative breast cancer in the general population. Nat Genet. 2010;42:885–92.
Chenevix-Trench G, Milne RL, Antoniou AC, Couch FJ, Easton DF, Goldgar DE, et al. An international initiative to identify genetic modifiers of cancer risk in BRCA1 and BRCA2 mutation carriers: the Consortium of Investigators of Modifiers of BRCA1 and BRCA2 (CIMBA). Breast Cancer Res. 2007;9:104.
Barnes DR, Rookus MA, McGuffog L, Leslie G, Mooij TM, Dennis J, et al. Polygenic risk scores and breast and epithelial ovarian cancer risks for carriers of BRCA1 and BRCA2 pathogenic variants. Genet Med. 2020;22:1653–66.
Couch FJ, Wang X, McGuffog L, Lee A, Olswold C, Kuchenbaecker KB, et al. Genome-wide association study in BRCA1 mutation carriers identifies novel loci associated with breast and ovarian cancer risk. PLoS Genet. 2013;9:e1003212.
Kuchenbaecker KB, McGuffog L, Barrowdale D, Lee A, Soucy P, Dennis J, et al. Evaluation of Polygenic Risk Scores for Breast and Ovarian Cancer Risk Prediction in BRCA1 and BRCA2 Mutation Carriers. J Natl Cancer Inst. 2017;109;djw302.
Mavaddat N, Michailidou K, Dennis J, Lush M, Fachal L, Lee A, et al. Polygenic Risk Scores for Prediction of Breast Cancer and Breast Cancer Subtypes. Am J Hum Genet. 2019;104:21–34.
Milne RL, Antoniou AC. Modifiers of breast and ovarian cancer risks for BRCA1 and BRCA2 mutation carriers. Endocr Relat Cancer. 2016;23:T69–84.
Milne RL, Kuchenbaecker KB, Michailidou K, Beesley J, Kar S, Lindstrom S, et al. Identification of ten variants associated with risk of estrogen-receptor-negative breast cancer. Nat Genet. 2017;49:1767–78.
Barnes DR, Rookus MA, McGuffog L, Leslie G, Mooij TM, Dennis J, et al. Polygenic risk scores and breast and epithelial ovarian cancer risks for carriers of BRCA1 and BRCA2 pathogenic variants. Genet Med. 2020;22:1653–66.
Bakhtiari M, Park J, Ding YC, Shleizer-Burko S, Neuhausen SL, Halldorsson BV, et al. Variable number tandem repeats mediate the expression of proximal genes. Nat Commun. 2021;12:2075.
Fotsing SF, Margoliash J, Wang C, Saini S, Yanicky R, Shleizer-Burko S, et al. The impact of short tandem repeat variation on gene expression. Nat Genet. 2019;51:1652–9.
Quilez J, Guilmatre A, Garg P, Highnam G, Gymrek M, Erlich Y, et al. Polymorphic tandem repeats within gene promoters act as modifiers of gene expression and DNA methylation in humans. Nucl Acids Res. 2016;44:3750–62.
Brookes KJ. The VNTR in complex disorders: the forgotten polymorphisms? A functional way forward? Genomics. 2013;101:273–81.
Gymrek M, Willems T, Guilmatre A, Zeng H, Markus B, Georgiev S, et al. Abundant contribution of short tandem repeats to gene expression variation in humans. Nat Genet. 2016;48:22–9.
Hofer P, Zochmeister C, Behm C, Brezina S, Baierl A, Doriguzzi A, et al. MNS16A tandem repeat minisatellite of human telomerase gene: functional studies in colorectal, lung and prostate cancer. Oncotarget. 2017;8:28021–7.
Rose AM, Krishan A, Chakarova CF, Moya L, Chambers SK, Hollands M, et al. MSR1 repeats modulate gene expression and affect risk of breast and prostate cancer. Ann Oncol. 2018;29:1292–303.
Krontiris TG, Devlin B, Karp DD, Robert NJ, Risch N. An association between the risk of cancer and mutations in the HRAS1 minisatellite locus. N Engl J Med. 1993;329:517–23.
Rajaei M, Saadat I, Omidvari S, Saadat M. Association between polymorphisms at promoters of XRCC5 and XRCC6 genes and risk of breast cancer. Med Oncol. 2014;31:885.
Wang Y, Hu Z, Liang J, Wang Z, Tang J, Wang S, et al. A tandem repeat of human telomerase reverse transcriptase (hTERT) and risk of breast cancer development and metastasis in Chinese women. Carcinogenesis. 2008;29:1197–201.
Xiang C, Gao H, Meng L, Qin Z, Ma R, Liu Y, et al. Functional variable number of tandem repeats variation in the promoter of proto-oncogene PTTG1IP is associated with risk of estrogen receptor-positive breast cancer. Cancer Sci. 2012;103:1121–8.
Bakhtiari M, Shleizer-Burko S, Gymrek M, Bansal V, Bafna V. Targeted genotyping of variable number tandem repeats with adVNTR. Genome Res. 2018;28:1709–19.
Antoniou AC, Spurdle AB, Sinilnikova OM, Healey S, Pooley KA, Schmutzler RK, et al. Common breast cancer-predisposition alleles are associated with breast cancer risk in BRCA1 and BRCA2 mutation carriers. Am J Hum Genet. 2008;82:937–48.
Coignard J, Lush M, Beesley J, O’Mara TA, Dennis J, Tyrer JP, et al. A case-only study to identify genetic modifiers of breast cancer risk for BRCA1/BRCA2 mutation carriers. Nat Commun. 2021;12:1078.
Kuchenbaecker KB, Neuhausen SL, Robson M, Barrowdale D, McGuffog L, Mulligan AM, et al. Associations of common breast cancer susceptibility alleles with risk of breast cancer subtypes in BRCA1 and BRCA2 mutation carriers. Breast Cancer Res. 2014;16:3416.
Rebbeck TR, Mitra N, Wan F, Sinilnikova OM, Healey S, McGuffog L, et al. Association of type and location of BRCA1 and BRCA2 mutations with risk of breast and ovarian cancer. JAMA 2015;313:1347–61.
Roa BB, Boyd AA, Volcik K, Richards CS. Ashkenazi Jewish population frequencies for common mutations in BRCA1 and BRCA2. Nat Genet. 1996;14:185–7.
Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucl acids Res. 1999;27:573–80.
Borgstrom E, Lundin S, Lundeberg J. Large scale library generation for high throughput sequencing. PLoS One. 2011;6:e19119.
Fisher S, Barry A, Abreu J, Minie B, Nolan J, Delorey TM, et al. A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol. 2011;12:R1.
Guo SW, Thompson EA. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics 1992;48:361–72.
Rebbeck TR, Kantoff PW, Krithivas K, Neuhausen S, Blackwood MA, Godwin AK, et al. Modification of BRCA1-associated breast cancer risk by the polymorphic androgen-receptor CAG repeat. Am J Hum Genet. 1999;64:1371–7.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc. 1995;57:289–300.
Barnes DR, Lee A, Investigators E, kConFab I, Easton DF, Antoniou AC. Evaluation of association methods for analysing modifiers of disease risk in carriers of high-risk mutations. Genet Epidemiol. 2012;36:274–91.
Therneau M, Grambsch P. Modeling Survival data: Extending the Cox Model. New York: Springer; 2000.
Clague J, Wilhoite G, Adamson A, Bailis A, Weitzel JN, Neuhausen SL. RAD51C germline mutations in breast and ovarian cancer cases from high-risk families. PLoS One. 2011;6:e25632.
Holm S. A simple sequentially rejective multiple test procedure. Scandanavian J Stat. 1979;6:65–70.
Zhang C, Lv GQ, Yu XM, Gu YL, Li JP, Du LF, et al. Current evidence on the relationship between HRAS1 polymorphism and breast cancer risk: a meta-analysis. Breast Cancer Res Treat. 2011;128:467–72.
Clark J, Smith SS. Secondary structure at a hot spot for DNA methylation in DNA from human breast cancers. Cancer Genomics Proteom. 2008;5:241–51.
Mao Q, Qiu M, Dong G, Xia W, Zhang S, Xu Y, et al. CAG repeat polymorphisms in the androgen receptor and breast cancer risk in women: a meta-analysis of 17 studies. Onco Targets Ther. 2015;8:2111–20.
Pu SY, Yu Q, Wu H, Jiang JJ, Chen XQ, He YH, et al. ERCC6L, a DNA helicase, is involved in cell proliferation and associated with survival and progress in breast and kidney cancers. Oncotarget. 2017;8:42116–24.
Liu J, Sun J, Zhang Q, Zeng Z. shRNA knockdown of DNA helicase ERCC6L expression inhibits human breast cancer growth. Mol Med Rep. 2018;18:3490–6.
Bahramian S, Sahebi R, Roohinejad Z, Delshad E, Javid N, Amini A, et al. Low expression of LncRNA-CAF attributed to the high expression of HIF1A in esophageal squamous cell carcinoma and gastric cancer patients. Mol Biol Rep. 2022;49:895–905.
Gilkes DM, Semenza GL. Role of hypoxia-inducible factors in breast cancer metastasis. Future Oncol. 2013;9:1623–36.
van der Groep P, Bouter A, Menko FH, van der Wall E, van Diest PJ. High frequency of HIF-1alpha overexpression in BRCA1 related breast cancer. Breast Cancer Res Treat. 2008;111:475–80.
van der Groep P, van Diest PJ, Smolders YH, Ausems MG, van der Luijt RB, Menko FH, et al. HIF-1alpha overexpression in ductal carcinoma in situ of the breast in BRCA1 and BRCA2 mutation carriers. PLoS One. 2013;8:e56055.
Lu D, Song J, Lu Y, Fall K, Chen X, Fang F, et al. A shared genetic contribution to breast cancer and schizophrenia. Nat Commun. 2020;11:4637.
Chang H, Xiao X, Li M. The schizophrenia risk gene ZNF804A: clinical associations, biological mechanisms and neuronal functions. Mol Psychiatry. 2017;22:944–53.
Klockmeier K, Silva Ramos E, Rasko T, Marti, Pastor A, Wanker EE. Schizophrenia risk candidate protein ZNF804A interacts with STAT2 and influences interferon-mediated gene transcription in mammalian cells. J Mol Biol. 2021;433:167184.
Lee HJ, Song JY, Kim JW, Jin SY, Hong MS, Park JK, et al. Association study of polymorphisms in synaptic vesicle-associated genes, SYN2 and CPLX2, with schizophrenia. Behav Brain Funct. 2005;1:15.
Li X, Su X, Liu J, Li H, Li M, Me Research T, et al. Transcriptome-wide association study identifies new susceptibility genes and pathways for depression. Transl Psychiatry. 2021;11:306.
Saviouk V, Moreau MP. Tereshchenko IV, Brzustowicz LM. Association of synapsin 2 with schizophrenia in families of Northern European ancestry. Schizophr Res. 2007;96:100–11.
Depienne C, Mandel JL. 30 years of repeat expansion disorders: What have we learned and what are the remaining challenges? Am J Hum Genet. 2021;108:764–85.
Antoniou AC, Hardy R, Walker L, Evans DG, Shenton A, Eeles R, et al. Predicting the likelihood of carrying a BRCA1 or BRCA2 mutation: validation of BOADICEA, BRCAPRO, IBIS, Myriad and the Manchester scoring system using data from UK genetics clinics. J Med Genet. 2008;45:425–31.
De Roeck A, Duchateau L, Van Dongen J, Cacace R, Bjerke M, Van den Bossche T, et al. An intronic VNTR affects splicing of ABCA7 and increases risk of Alzheimer’s disease. Acta Neuropathol. 2018;135:827–37.
Liu G, Li F, Zhang S, Jiang Y, Ma G, Shang H, et al. Analyzing large-scale samples confirms the association between the ABCA7 rs3764650 polymorphism and Alzheimer’s disease susceptibility. Mol Neurobiol. 2014;50:757–64.
Song JHT, Lowe CB, Kingsley DM. Characterization of a Human-Specific Tandem Repeat Associated with Bipolar Disorder and Schizophrenia. Am J Hum Genet. 2018;103:421–30.
Acknowledgements
We thank the women for participating in the study.
Funding
This work was supported by the Basser Foundation (SLN and EF) and the Beckman Research Institute of City of Hope Excellence Awards Program. Sequencing of the VNTRs was performed in the Integrative Genomics Core supported by the National Cancer Institute of the National Institutes of Health under grant number P30CA033572. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. SLN is partially supported by the Morris and Horowitz Families Professorship. VB and JP were supported in part by R01GM114362, R01HG010149 and RO1HG011558 from the NIH.
Author information
Authors and Affiliations
Contributions
SLN and EF conceived of the idea, developed the design, and obtained funding to conduct the study. VB, MB, and JP did the analysis of the sequencing data to obtain genotypes. AA and CP prepared the DNA libraries for sequencing, conducted the luciferase assays, and the validation of the VNTR genotypes. YCD did quality control of the VNTR genotypes and conducted the association statistical analysis. SLN, EF, JW, and YL contributed DNA samples and data. All authors have contributed to, read, and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
V.Bafna is a co-founder, paid consultant, SAB member and has equity interest in Boundless Bio, inc. and Abterra, Inc. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies. None of the other authors have any potential competing interests.
Ethics approval and consent to participate
All individuals provided voluntary, informed consent. Ethics approvals were through each center-specific Institutional Review Board approved protocols.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ding, Y.C., Adamson, A.W., Bakhtiari, M. et al. Variable number tandem repeats (VNTRs) as modifiers of breast cancer risk in carriers of BRCA1 185delAG. Eur J Hum Genet 31, 216–222 (2023). https://doi.org/10.1038/s41431-022-01238-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-022-01238-z
This article is cited by
-
The value of exomes across the ages
European Journal of Human Genetics (2023)