Abstract
Most secondary genomic findings (SFs) fall in the scope of primary care practice. However, primary care providers' (PCPs) capacity to manage these findings is not well understood. We explored PCPs’ views and experiences of managing SFs through a qualitative study. PCPs participated in semi-structured interviews about SFs from a patient in their practice or a hypothetical patient. The interpretive descriptive methodology was used to analyze transcripts thematically through constant comparison. Fifteen family physicians from Ontario, Canada participated (ten females; 6–40 years in practice across community and academic settings). PCPs made sense of SFs through the lens of actionability: they actively looked for clinical relevance by considering a wide range of immediate and future actions, including referrals, genetic testing, screening, lifestyle changes, counseling about family planning, informing family members, future medication choice, increased vigilance/surveillance, and managing results in the electronic medical record. PCPs saw clinical actionability as the main benefit mitigating the potential harms of learning SFs, namely patient anxiety and unnecessary investigations. PCPs conceptualized actionability more broadly than it is traditionally defined in medical genetics. Further research will be needed to determine if PCPs’ emphasis on actionability conflicts with patients’ expectations of SFs and if it leads to overutilization of healthcare resources.
Similar content being viewed by others
Introduction
Genomic sequencing (GS), encompassing exome and genome sequencing, increasingly is available in clinical practice. There is growing evidence that GS improves diagnostic yield for hereditary diseases in a variety of settings [1,2,3]. GS may also reveal incidental findings to the primary reason for testing, such as medically actionable results for which direct treatment or prevention is available, pharmacogenomic variants, common disease single nucleotide polymorphisms (SNPs) conferring small risk changes, results for rare, Mendelian genetic diseases, and carrier status results [4, 5]. We use secondary findings (SFs) to include all clinically significant results in addition to those deemed medically actionable by the American college of medical genetics and genomics (ACMG) [6]. Current guidance varies on whether SFs should be reported: the ACMG suggests a return of medically actionable results with patient consent while the European society of human genetics (ESHG) recommends against opportunistic genomic screening and routine return of SFs outside of research settings [6, 7]. Yet, patients are increasingly interested in learning SFs beyond those that are medically actionable [8,9,10].
Cohort studies indicate that over 85% of SFs generated from GS are common disease, pharmacogenomics, or carrier status results, which may largely fall into the scope of primary care [11]. Primary care providers (PCPs) are also likely to be tasked with managing SFs because of the shortage of genetics specialists, especially in rural areas: there are only about two geneticists or genetic counselors per 100,000 Americans, 1 per 100,000 Canadians, and fewer than 0.5–1.2 per 100,000 Europeans [12,13,14]. However, PCPs describe barriers to managing conventional genetic tests such as limited knowledge, time, and access to genetics specialists [15,16,17]. These barriers are likely magnified in dealing with the volume and variety of SFs. PCPs are crucial to SF management in clinical care, but literature exploring their capacity to manage these findings is very limited. Our objective was to describe PCPs’ experiences and views of managing SFs from GS, an important step in the anticipated large-scale adoption of genomic medicine.
Materials and methods
Design
Qualitative methods, specifically interpretive descriptive methodology, were used to explore PCPs’ experiences, meanings, and motivations in managing SFs [18, 19]. This was a sub-study of an ongoing randomized controlled trial (NCT03597165) recruiting patients with suspected hereditary cancer to receive SFs from exome sequencing; the trial protocol is described elsewhere [20]. In brief, patients were randomized to a control arm (receiving only variants associated with cancer) or an intervention arm (receiving cancer results and SFs of their choice). Intervention arm patients chose to receive SFs from any combination of five categories based on a validated decision-aid developed through a usability study: (1) medically actionable and pharmacogenomic results; (2) common disease SNPs; (3) rare Mendelian results; (4) early-onset neurological results; and (5) carrier status results [5, 21]. Table 1 provides examples of the SFs in the actual and hypothetical reports. Patients received reports developed by laboratory and medical geneticists and genetic counselors in which results were generated through a comprehensive pipeline and reported in the five categories above [20, 22]. The Unity Health Toronto Research Ethics Board approved this qualitative study in January 2020.
Participants and recruitment
PCPs were recruited in two groups using courier mail, email, telephone, and fax. Group 1 PCPs had a patient in their practice in the trial intervention arm. All potential group 1 participants were invited. Group 2 PCPs were family physicians recruited from the professional networks of JCC, YB, and MV through maximal variation sampling across sex, geographic setting, and academic setting [23]. Since they were outside the trial, group 2 PCPs were all provided the same sample sequencing report and consultation summary letter representative of reports and letters in the trial. Prior to recruitment, no provider had a direct relationship to the study. Participants were recruited until data saturation was achieved, i.e., new information or themes were no longer emerging from the data [23].
Data collection
Individual semi-structured phone or in-person interviews were conducted; field notes were taken concurrently. AS conducted all interviews, including three pilot interviews to refine the interview guide and train the interviewer. The interview guide covered the PCPs’ actual or hypothetical experiences in managing the SFs, impact on clinical management, and prior genetics knowledge and experience. The guide was informed by the literature review and pilot interviews and iteratively adapted over the course of data collection (see Appendix 1). Interviews were audio-recorded and transcribed.
Data analysis
Data analysis was iterative with data collection [19]. As per the interpretive description, data analysis began with repeated immersion in the data by reading and listening to each interview multiple times. Initial codes were then identified inductively from the first five interviews to capture larger, more meaningful sections of data [19, 23]. Linkages and themes were created by constant comparison within and between interviews by AS, YB, JCC, and MV. Dedoose, a web-based qualitative software, was used to manage codes.
Results
Participants’ characteristics
Fifteen family physicians practicing in Ontario, Canada participated in interviews of about 50 min (range: 17–66 min). Three were from group 1 (PCPs with a patient in the trial and received a real SF report and consult letter), and 12 were from group 2 (received the hypothetical SF report and letter to review). All participants were in a group practice with variation across age, years of practice, and practice setting (Table 2). Most participants were female (10/15) and were urban or suburban practitioners (12/15). The results received by group 1 were similar to those presented to group 2: all reports and letters included common disease SNPs, carrier status and pharmacogenomic results, and either negative or uncertain cancer results. However, the patients of the group 1 PCPs did not happen to have any medically actionable results as defined by the ACMG.
PCPs made sense of the SFs through the lens of actionability
While PCPs displayed a range in attitudes towards SFs from “interesting information” [GP1-2] to a “nightmare” [GP2-4], PCPs in both groups sought clinical relevance and concrete actions they could take once they learned their patients’ SFs:
“Now you have this information, and what do you do with it? Like is it just an intellectual exercise or is there something concrete that you can do with this knowledge?” [GP2-5]
PCPs saw actions as a way to use the results to have a “real impact on my patients” [GP2-8] to potentially minimize the risk of disease and improve outcomes. PCPs also perceived a medico-legal risk and responsibility to the patient to act on known information appropriately:
“Is there any medical liability because I didn’t follow up closer for something that there was a possible risk…now that you know something, you definitely need to act on it and do something about it one way or another.” [GP2-1]
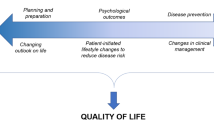
Thus, PCPs in both groups approached SFs through the lens of actionability: by looking for clinical actions that could be taken based on this information (Fig. 1).
Actionability was the main benefit to learning SFs
PCPs saw actionability as the main benefit of learning any health information, including SFs. However, they did not consider all SFs to be beneficial because they did not perceive all SFs to be actionable.
All PCPs saw medically actionable and pharmacogenomic results as leading to clear actions that could benefit their patient’s current or future health:
“You can actually do something…it could lead to severe health problems if it’s not picked up early. But there’s a treatment available, so that to me is the ideal thing being found as an incidental finding.” [GP2-9]
The actions they described included referrals, alternative medications or dosages, and entering this information prominently into the electronic medical record (EMR) for future clinical decision making. Thus, all PCPs saw the benefit of medically actionable and pharmacogenomic SFs.
However, PCPs varied in whether they identified a potential course of action and thus benefit for learning common disease, early-onset neurological, and carrier status results. For example, PCPs did not consistently see the SFs for age-related macular degeneration as beneficial because some PCPs did not consider or were not aware of preventive actions.
“But, if there’s a result that I can’t do anything about then I wonder what’s the value of that result. Except, as you said, like it might inform so you just say ‘hey, just so you know, you might be at increased risk for macular degeneration down the road. So, make sure you have a well-lit house.’ Because like what else can you do for macular degeneration? Like not a lot.” [GP1-1]
In contrast, other PCPs considered preventive actions to be possible for age-related macular degeneration and thus saw the benefit of learning this result.
“First of all, there’s a lot of lifestyle things that we can do for the macular degeneration. Protect the eyes from sunshine, don’t smoke, eat lots of dark green leafy vegetables, and…you can refer them earlier rather than later to an ophthalmologist for assessment in that they could catch the disease early and hopefully slow it down or reverse it.” [GP2-9]
PCPs similarly varied in their perception of the actionability of carrier status results. A minority of PCPs did not think carrier status results could lead to any actions, referring to them as “a dead-end.” [GP1-1]. However, most PCPs saw some benefit to carrier status results because they saw value in actions such as counseling, carrier testing, and referrals. Since these actions might not directly affect the patient unless they were thinking of having children, PCPs managed carrier status in a variety of ways. Some left it to their patients to prompt action, while others considered informing family members in their practice with the patient’s permission. However, a few PCPs grappled with whether they were responsible to act on the carrier result by helping the patient to inform their family: one participant described how if their patient was not “left with the kind of appropriate way maybe to relay that or whatever, it may come to me to actually provide the support that she might need to…manage that.” [GP2-11]
PCPs had a wide lens of actionability
PCPs considered a wide range of immediate and future actions based on the SFs (Table 3). Among immediate actions, many first considered whether to initiate a patient visit because they anticipated that the patient would be overwhelmed and would want to discuss the results. Others felt a consultation was unnecessary if the findings were not applicable to their patient’s immediate health status. PCPs also assessed whether any immediate clinical actions needed to be taken beyond a patient visit, such as referrals, family genetic testing, screening, and discussing lifestyle changes:
“If there’s an action to be taken or if there’s things that we can do like for example…let’s say they could really focus on diet and lifestyle, and quitting smoking and cutting out alcohol would reduce their chances.” [GP2-5]
Most PCPs also considered “counseling about family planning” [GP2-7] and informing family members among immediate clinical actions.
In addition, PCPs considered future clinical actions that could arise from the SFs, such as medication choice based on pharmacogenomic results, or increased vigilance and earlier investigation if their patient presented in the future with non-specific symptoms consistent with a disease for which they were at genetic risk:
“Maybe they have rectal bleeding…If I was to flick up there [to the patient’s chart] and be like ‘oh there’s this increased risk of Crohn’s Disease.’ It might tip my decision-making or I’d have that, it would be part of my note.” [GP2-2]
To enable these future actions, PCPs suggested placing the information in a prominent place in their EMRs. In this way, information management was itself considered a possible action resulting from the SFs.
“That’s something that might affect her care down the road. So, my responsibility as the family doctor is to keep track of that information so it doesn’t get lost. So, that is an actionable outcome.” [GP1-1]
PCPs were selective about the information they chose to store prominently in their EMRs, limiting it to results that could lead to actions they were responsible to initiate:
“I would only put things in the…CPP [cumulative patient profile]…where I might be starting to do something that I need to…you know…actually inform that; so drugs, screening…ophthalmology or optometry exams.” [GP2-11]
Actionability mitigated the potential harms of learning SFs
To many PCPs, “information without some action that can be done at the end of the day is useless and potentially harmful” [GP1-1]. Even those PCPs who stated that “knowledge is power” [GP1-2] clarified that knowledge needed to change management in some way; otherwise, it could lead to harm. Therefore, without actionability, PCPs described that patients were only left with the potential harms of learning SFs.
The main harm concerning to PCPs was the potential for “anxiety in people especially if they know that they can’t do anything” [GP2-7]. This was particularly apparent when considering early-onset neurological disease results. Since most PCPs felt little could be done about these results, they described how patients could perceive the result as a certainty like a “ticking time bomb” [GP1-2] and experience anxiety. While PCPs acknowledged that not everyone may react to SFs with anxiety, the rising general and health anxiety they observed in primary care practice heightened their concern about these outcomes. They stated that this anxiety would fall on them to manage despite their lack of familiarity with genomics and its ramifications.
The other harm raised by PCPs was the potential for unnecessary follow-up investigations with physical and psychological patient harm. PCPs were uncertain in some cases whether there was evidence to justify follow-up based on SFs. This came up most when considering changes to screening based on common disease results. Without a clear “cut off for the odds ratio that would require more close surveillance” [GP2-7], PCPs described how patients with an SF for increased risk of Crohn’s disease, for example, might push for unwarranted further investigation such as a colonoscopy that carries serious physical risk. In the PCPs’ experience, even follow-up investigations that carried less risk, such as blood tests or imaging, could have a negative “psychological impact” [GP2-12] on patients. PCPs were especially concerned because patients often did not realize the potential harms that could stem from follow-up investigations.
“They’re like ‘oh, what’s the harm in finding out?’ Like they just think they’re getting more information but they don’t realize what that information can then lead to and what the potential harms can arise from that.” [GP2-1]
Some of their concern came from the “escalating costs of healthcare” [GP1-1] and their perceived obligation to be stewards of healthcare resources. PCPs mentioned campaigns such as Choosing Wisely Canada, which encourages the reduction of unnecessary tests.
“That’s the whole Choosing Wisely paradigm now. Right? Our resource stewardship is terrible. We order stuff all the time that we shouldn’t be ordering and then we find all sorts of random stuff.” [GP2-12]
PCPs questioned the benefit of information that was not actionable, though they acknowledged some patients wanted to learn as much health information as possible:
“Is it going to be helpful or harmful? And, just because a patient says ‘I want to know everything,’ I don’t know that…unless the advantages outweigh the disadvantages, I don’t know that it makes sense to do it.” [GP2-4]
When prompted to discuss why patients might want to know all information, many PCPs believed it was a “philosophical thing…some patients just want to know.” [GP2-3] Other PCPs felt that patients might assume that everything was actionable.
I think people often assume that if I find this information out, finding it out will make a difference…I do want to emphasize I don’t think we should never do this. But I’m talking specifically about the cases where…there’s no action that comes from the information other than just now you know it to be true” [GP2-5].
However, no PCPs would withhold non-actionable results from their patients because they did not consider themselves the “gatekeeper” of information [GP2-3]. Therefore, PCPs wanted to respect their patient’s right to know their own health information but felt patients may not gain anything from SFs that were not actionable.
Discussion
Despite the potential increase in SFs that are returned and fall into the scope of primary care practice, little is known about how PCPs approach the management of these results. Our study is the first to describe that PCPs make sense of SFs through the lens of actionability: by looking for clinical actions that could be taken once this information was known to them. Actions were the way in which PCPs could use the results to make a difference to their patient’s health. PCPs conceptualized actionability as a wide range of possible clinical actions, both those that could occur immediately (e.g., referrals and discussion of lifestyle changes) and those that could become relevant in the future (e.g., information management to enable future treatment). PCPs saw actionability as the main benefit to learning SFs, mitigating potential harms to patients and the healthcare system, such as patient anxiety and possible unnecessary investigations.
The study’s results revealed three major tensions. The first tension is about overuse. PCPs perceive a responsibility to reduce investigations with minimal or uncertain benefit, mainly unnecessary tests, which may be in tension with the wide range of actions they considered possible from SFs [24,25,26]. Unnecessary costs may arise if, after receiving SFs, the actions considered by PCPs and requested by patients are not all based on evidence or best practices. One study observed that PCPs recommended twice as many clinical actions when SFs were available compared to when family history alone was available, concluding that these actions were of potentially unclear value [11]. However, long-term savings could arise from preventive care actions considered by PCPs (although genetic testing has not been shown to motivate lifestyle changes to date) [27]. Thus, further research, especially pilot trials, is needed to investigate long-term outcomes, resource use, and cost-effectiveness in managing SFs in primary care [28].
The second tension is between PCPs’ and patients’ values regarding SFs. While PCPs focused on actionability, the literature shows that many patients desire access to their genomic information regardless of clinical actionability [9, 10, 29]. For patients, other benefits to learning SFs were emotional/financial planning for future disease, alleviation of uncertainty, control gained by learning health information, and growing value of SFs as science advances. Patients’ primary result, diagnosis, life stage, and personal values also influenced their perception of these benefits [9, 29]. The divergence in perspective between patients and PCPs could affect practice since PCPs may not initiate discussion of findings that they do not consider actionable. As in the case of age-related macular degeneration in our study, PCPs may vary in whether they see a finding as actionable. To bridge this gap and promote care equity, shared decision-making could help attune PCPs to patients’ values when deciding which SFs to receive and manage [30]. While shared decision-making is unlikely to limit the information patients want, it could allow PCPs to discuss harms their patients may not have considered and leverage their relationship with their patients to facilitate decision-making [31, 32]. Thus, shared decision-making may help lessen the tension between PCPs’ and patients’ values regarding SFs.
The third tension involves their role in medical care. PCPs conceptualized actionability beyond its traditional definition in medical genetics, which is typically referred to as medically actionable results with established interventions. PCPs’ widened sense of actionability aligns with Starfield’s four essential functions of primary care: first-contact care, continuous care, comprehensive care, and coordinated care [33]. The functions show that, unlike in medical genetics, PCPs are responsible for providing comprehensive care over a patient’s lifetime including coordinating with specialists through referrals and follow-up care across many countries in Europe and Canada [33]. Thus, based on their role, it is appropriate that PCPs conceptualize care and actions more broadly than medical genetics professionals. Furthermore, PCPs feel they may need to take some action if receiving SFs, even if those SFs are not always judged actionable in medical genetics. In fact, PCPs will be responsible for managing the pharmacogenomic variants, common disease SNPs, and/or carrier status results that 85–100% of patients could receive through GS [11]. Therefore, a difference exists between the roles of primary care and medical genetics in their approach to managing SFs.
These tensions between patients, providers, and disciplines parallel the ongoing debate about the clinical and personal utility of SFs [34, 35]. A narrower approach to SFs that defines utility as strictly clinical limits harms but may miss potential benefits, such as the cautious approach of the ESHG, the 59 genes considered medically actionable by the ACMG, or the 14 genes considered by Genomics England. In contrast, a broader approach encompassing SFs that offer clinical and personal utility allows for social benefits while potentially accepting harms. To reconcile these tensions, further research must deeply explore the potential harms and benefits of SFs, including how PCPs’ actions may change the harm/benefit equation.
Limitations
There are several limitations to this study. First, despite efforts to recruit as many group 1 participants as possible, group 2 PCPs predominated the sample and it is possible their hypothetical experiences may not reflect PCPs’ actual experiences when experiencing the strains of busy practice [36]. However, we observed a consistent approach to managing SFs between groups 1 and 2 in this study. There is potential for selection bias in non-random sampling; however, we did not purposefully sample for PCPs with a strong interest in genetics. While results were limited in capturing the perspective of rural PCPs, our sample reflects diverse practice characteristics considering the breadth in age, years in practice, and setting across the hospital, university/academic, and community practice. In addition, this study did not explore result validity: whether participants would have acted differently if they were more aware that the SFs were research results and needed to be clinically confirmed. Finally, the sequencing results differed slightly between groups. Group 1 PCPs spoke about the results of their actual patient, which varied naturalistically. All group 2 PCPs received the same results about a hypothetical patient with breast cancer who received negative cancer results but who had medically actionable, pharmacogenomic, common disease, and carrier status SFs. Nevertheless, group 2 PCPs’ sequencing results closely reflected those of a typical trial participant.
Conclusion
PCPs approached SFs through the lens of actionability because they view actions as the way in which medical information can change patient health. PCPs have a wider sense of actionability than traditionally defined in clinical genetics because they considered a wide range of possible immediate and future clinical actions to address the results. PCPs saw actionability as a benefit mitigating the potential harms of learning SFs. This study suggests that PCPs’ concept of actionability may have some tension with what patients value about SFs, with their own perceived obligation towards medical resource stewardship, and may vary from that of the traditional role of clinical genetics. Further research is needed to explore these tensions and whether some may be addressed by shared decision-making.
References
Walsh M, Bell KM, Chong B, Creed E, Brett GR, Pope K, et al. Diagnostic and cost utility of whole exome sequencing in peripheral neuropathy. Ann Clin Transl Neurol. 2017;4:318–25.
Retterer K, Juusola J, Cho MT, Vitazka P, Millan F, Gibellini F, et al. Clinical application of whole-exome sequencing across clinical indications. Genet Med. 2016;18:696–704.
Groopman EE, Marasa M, Cameron-Christie S, Petrovski S, Aggarwal VS, Milo-Rasouly H, et al. Diagnostic utility of exome sequencing for kidney disease. N Engl J Med. 2019;380:142–51.
Berg JS, Khoury MJ, Evans JP. Deploying whole genome sequencing in clinical practice and public health: Meeting the challenge one bin at a time. Genet Med. 2011;13:499–504.
Bombard Y, Clausen M, Mighton C, Carlsson L, Casalino S, Glogowski E, et al. The genomics ADvISER: development and usability testing of a decision aid for the selection of incidental sequencing results. Eur J Hum Genet. 2018;26:984–95.
Kalia SS, Adelman K, Bale S, Chung WK, Eng C, Evans J, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American college of medical genetics and genomics. Genet Med. 2017;19:249–55.
Wert G De, Dondorp W, Clarke A, Dequeker EMC, Cordier C, Deans Z et al. Opportunistic genomic screening. Recommendations of the European Society of Human Genetics. Eur J Hum Genet. 2020;29:365–77.
Regier DA, Peacock SJ, Pataky R, Van Der Hoek K, Jarvik GP, Hoch J, et al. Societal preferences for the return of incidental findings from clinical genomic sequencing: a discrete-choice experiment. CMAJ. 2015;187:E190–97.
Clift KE, Halverson CME, Fiksdal AS, Kumbamu A, Sharp RR, McCormick JB. Patients’ views on incidental findings from clinical exome sequencing. Appl Transl Genom. 2015;4:38–43.
Ploug T, Holm S. Clinical genome sequencing and population preferences for information about ‘incidental’ findings—from medically actionable genes (MAGs) to patient actionable genes (PAGs). PLoS ONE. 2017;12:e0179935.
Vassy JL, Christensen KD, Schonman E, Blout C, Robinson JO, Krier JB, et al. The impact of whole-genome sequencing on the primary care and outcomes of healthy adult patients: a pilot randomized trial. Ann Intern Med. 2017;167:159–69.
Berberich AJ, Ho R, Hegele RA. Whole genome sequencing in the clinic: empowerment or too much information? CMAJ. 2018;190:E124–E125.
Maiese DR, Keehn A, Lyon M, Flannery D, Watson M. Current conditions in medical genetics practice. Genet Med. 2019;21:1874–1877.
Dragojlovic N, Borle K, Kopac N, Ellis U, Birch P, Adam S et al. The composition and capacity of the clinical genetics workforce in high-income countries: a scoping review. Genet Med. 2020;22:1437–49.
Mikat-Stevens NA, Larson IA, Tarini BA. Primary-care providers’ perceived barriers to integration of genetics services: a systematic review of the literature. Genet Med. 2015;17:169–76.
Carroll JC, Allanson J, Morrison S, Miller FA, Wilson BJ, Permaul JA et al. Informing integration of genomic medicine into primary care: an assessment of current practice, attitudes, and desired resources. Front Genet. 2019;10:1189.
Carroll JC, Makuzawa T, Manca DP, Sopcak N, Permaul JA, O’Brien MA, et al. Primary care providers’ experiences with and perceptions of personalized genomic medicine. Can Fam Phys. 2016;62:626–35.
Pope C, Mays N. Reaching the parts other methods cannot reach: an introduction to qualitative methods in health and health services research. BMJ. 1995;311:42–45.
Thorne S, Reimer Kirkham S, MacDonald-Emes J. Focus on qualitative methods. Interpretative description: a noncategorical qualitative alternative for developing nursing knowledge. Res Nurs Heal. 1997;20:169–77.
Shickh S, Clausen M, Mighton C, Gutierrez Salazar M, Zakoor KR, Kodida R, et al. Health outcomes, utility and costs of returning incidental results from genomic sequencing in a Canadian cancer population: protocol for a mixed-methods randomised controlled trial. BMJ Open. 2019;9:e031092.
Bombard Y, Clausen M, Shickh S, Mighton C, Casalino S, Kim THM, et al. Effectiveness of the genomics ADvISER decision aid for the selection of secondary findings from genomic sequencing: a randomized clinical trial. Genet Med. 2020;22:727–35.
Reble E, Gutierrez Salazar M, Zakoor K-R, Khalouei S, Clausen M, Kodida R et al. Beyond medically actionable results: an analytical pipeline for decreasing the burden of returning all clinically significant secondary findings. Hum Genet. 2020;140:493–504.
Thorne S. Interpretive description: qualitative research for applied practice. 2nd ed. New York; London: Routledge: Taylor & Francis. 2016.
Carroll JC, Grad R, Allanson JE, Pluye P, Permaul JA, Pimlott N, et al. The gene messenger impact project: an innovative genetics continuing education strategy for primary care providers. J Contin Educ Health Prof. 2016;36:178–85.
Riggs KR, Knight SJ. The language of stewardship: is the “low-value” label overused?. Mayo Clin Proc. 2017;92:11–14.
Wolfson DB, Tucker L. Foundations supporting stewardship of health care resources through medical education and training. Heal Aff Blog. 2014.
Hollands GJ, French DP, Griffin SJ, Prevost AT, Sutton S, King S et al. The impact of communicating genetic risks of disease on riskreducing health behaviour: systematic review with meta-analysis. BMJ. 2016;352:i1102.
Stark Z, Dolman L, Manolio TA, Ozenberger B, Hill SL, Caulfied MJ, et al. Integrating genomics into healthcare: a global responsibility. Am J Hum Genet 2019;104:13–20.
Mighton C, Carlsson L, Clausen M, Casalino S, Shickh S, McCuaig L, et al. Development of patient “profiles” to tailor counseling for incidental genomic sequencing results. Eur J Hum Genet. 2019;27:1008–17.
Vanstone M, Kinsella AE, Nisker J. Information-sharing to promote informed choice in prenatal screening in the spirit of the SOGC clinical practice guideline: a proposal for an alternative model. J Obstet Gynaecol Can. 2012;34:269–75.
Sanderson SC, Linderman MD, Suckiel SA, Diaz GA, Zinberg RE, Ferryman K, et al. Motivations, concerns and preferences of personal genome sequencing research participants: baseline findings from the HealthSeq project. Eur J Hum Genet. 2015;24:1–7.
Cunniff C, Bassetti J. Advances in genetic medicine and shared-decision making. J Commun Health. 2019;12:82–85.
Starfield B, Holtzman NA, Roland MO, Sibbald B, Harris R, Harris H. Primary care and genetic services: Health care in evolution. Eur J Public Health. 2012;12:51–56.
Mighton C, Carlsson L, Clausen M, Casalino S, Shickh S, McCuaig L et al. Quality of life drives patients’ preferences for secondary findings from genomic sequencing. Eur J Hum Genet. 2020;28:1178–86.
Kohler JN, Turbitt E, Biesecker BB. Personal utility in genomic testing: a systematic literature review. Eur J Hum Genet. 2017;25:662–8.
Ormondroyd E, Harper AR, Thomson KL, Mackley MP, Martin J, Penkett CJ, et al. Secondary findings in inherited heart conditions: a genotype-first feasibility study to assess phenotype, behavioural and psychosocial outcomes. Eur J Hum Genet. 2020;28:1486–96.
Acknowledgements
We would like to thank Dr. Ross Upshur and Dr. Holly Etchegary for their insight and feedback with respect to this study.
Funding
This project is partially funded by an Early Career Award from the Ontario Ministry of Research and Innovation (ER17-13-045). YB was supported by a Canadian Institutes of Health Research New Investigator Award and a Foundation Grant (FRN#143310). AS was supported by an Ontario Graduate Scholarship from the University of Toronto.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Sebastian, A., Carroll, J.C., Vanstone, M. et al. Widening the lens of actionability: A qualitative study of primary care providers’ views and experiences of managing secondary genomic findings. Eur J Hum Genet 30, 595–603 (2022). https://doi.org/10.1038/s41431-021-00876-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-021-00876-z
This article is cited by
-
Parents’ attitudes towards research involving genome sequencing of their healthy children: a qualitative study
European Journal of Human Genetics (2024)
-
“I just wanted more”: Hereditary cancer syndromes patients’ perspectives on the utility of circulating tumour DNA testing for cancer screening
European Journal of Human Genetics (2024)
-
No gene to predict the future?
European Journal of Human Genetics (2022)
-
A comprehensive genomic reporting structure for communicating all clinically significant primary and secondary findings
Human Genetics (2022)