Abstract
Hirschsprung disease (HSCR) is a congenital disorder with a population incidence of ~1/5000 live births, defined by an absence of enteric ganglia along variable lengths of the colon. HSCR genome-wide association studies (GWAS) have found common associated variants at RET, SEMA3, and NRG1, but they still fail to explain all of its heritability. To enhance gene discovery, we performed a GWAS of 170 cases identified from the Danish nationwide pathology registry with 4717 controls, based on 6.2 million variants imputed from the haplotype reference consortium panel. We found a novel low-frequency variant (rs144432435), which, when conditioning on the lead RET single-nucleotide polymorphism (SNP), was of genome-wide significance in the discovery analysis. This conditional association signal was replicated in a Swedish HSCR cohort with discovery plus replication meta-analysis conditional odds ratio of 6.6 (P = 7.7 × 10−10; 322 cases and 4893 controls). The conditional signal was, however, not replicated in two HSCR cohorts from USA and Finland, leading to the hypothesis that rs144432435 tags a rare haplotype present in Denmark and Sweden. Using the genome-wide complex trait analysis method, we estimated the SNP heritability of HSCR to be 88%, close to estimates based on classical family studies. Moreover, by using Lasso (least absolute shrinkage and selection operator) regression we were able to construct a genetic HSCR predictor with a area under the receiver operator characteristics curve of 76% in an independent validation set. In conclusion, we combined the largest collection of sporadic Hirschsprung cases to date (586 cases) to further elucidate HSCR’s genetic architecture.
Similar content being viewed by others
Introduction
Hirschsprung disease (HSCR: MIM 142623), or aganglionic megacolon, is a congenital disorder defined by an absence of enteric ganglia along variable lengths of the large intestine [1]. It has a population incidence of ~1/5000 live births, a sex ratio of 4:1 male:female, and a high heritability (>80%) with sibling recurrence risk 250-fold greater than that in the general population [1, 2]. HSCR is usually classified as short-segment HSCR (S-HSCR, 80% of HSCR cases), if aganglionosis goes no further than the upper sigmoid colon; as long-segment HSCR (L-HSCR, 15–20% of HSCR cases), if aganglionosis continues up to the splenic flexure and beyond; or as total colonic aganglionosis (TCA, ∼5% of HSCR cases), if aganglionosis affects the entire large intestine [1]. The condition results from failure in the migration, survival, proliferation, or differentiation of enteric neural crest-derived cells in the intestinal tract [3]. HSCR is one of the most common causes of intestinal obstruction in neonates [1, 4], with typical presentation of the disease being a newborn without bowel movement or a young child with severe chronic constipation. Treatment is surgical resection of the aganglionic colon [5]. HSCR is most frequently seen as isolated and non-syndromic cases (~80%), although familial aggregation and co-occurrence with chromosomal anomalies, such as Down syndrome, or other syndromes, have been documented [6]. HSCR is named after the Danish pediatrician Dr. Harald Hirschsprung, who first described it in 1888 [7], but despite having been known for more than a century, the etiology of the disease is not fully understood. The genetic architecture of HSCR is multifactorial and heterogeneous, influenced by both common and rare variants [6, 8]. Rare high-penetrance mutations have been found in genes associated with the development of the enteric nervous system, but they cumulatively explain <10% of cases [9]. Recent HSCR genome-wide association studies (GWAS) have found significantly greater phenotypic variation in common associated variants at RET, SEMA3, and NRG1 loci, but they still fail to explain all of its heritability [9,10,11,12,13]. Therefore, it is likely that additional common or low-frequency genetic variants associated with isolated HSCR are yet to be found. Our aim was to do a GWAS on all isolated HSCR cases available in the Danish National Biobank in order to unravel possible novel genetic loci involved in the disease. We performed a GWAS on 170 cases with a pathology-confirmed diagnosis of HSCR, and 4717 controls from other projects, all genotyped on the Illumina HumanOmniExpressExome array.
Materials and methods
Study subjects
Cases were defined as children with a pathology-confirmed diagnosis of aganglionosis as given by occurrence of the SNOMED-code S62590 (Mb. Hirschsprung) in the Danish National Pathology Registry [14]. They also had to be singletons born in Denmark with parents and grandparents born in Northwestern Europe (birthplace taken from the Danish Civil Registration System [15]). The cases were also required to not have any chromosomal anomalies or congenital malformations affecting multiple systems. Controls were defined as samples genotyped on the same array as the cases from previous GWAS projects at Statens Serum Institut, namely studies on febrile seizures [16] (n = 2035), hypospadias [17] (n = 1029), hydrocephalus (n = 766), and post-partum depression (n = 1113). All samples from the discovery stage were drawn from the Danish Neonatal Screening Biobank (part of the Danish National Biobank), which includes dried blood spot samples from babies born in Denmark from 1982 and onwards [18]. In the discovery stage, we started with 179 cases and 4943 controls, while in the replication stage we had three cohorts: 189 cases and 426 controls from USA, 152 cases and 176 controls from Sweden, and 75 cases and 301 controls from Finland, all of European ancestry. The Danish Scientific Research Ethics Committee of the Capital Region (Copenhagen) and the Danish Data Protection Agency approved this study. The Danish Scientific Ethics Committee also granted exemption from obtaining informed consent from participants as this research project was based on genotyping samples from biobank material (H-4-2013-055). For the replication cohorts, the Ethics Committees at the Karolinska Institutet and University of Helsinki approved the use of the Swedish and Finnish samples, respectively, while the institutional Committee on Human Research at Johns Hopkins University School of Medicine approved the use of samples from the USA.
Genotyping and quality control
All samples from the discovery stage were drawn from the Danish Neonatal Screening Biobank, part of the Danish National Biobank, hosted at Statens Serum Institut, Copenhagen, Denmark. The DNA from these samples was extracted and whole genome amplified from punches of dried blood spots using a previously described protocol [19]. The samples were then genotyped with the Illumina HumanOmniExpressExome array. Stringent quality control criteria were applied to the samples and variants in order to sequentially remove: (1) variant probes not aligned to the genome or that had >1 high-quality match (>90%) to the genome, (2) variants not present in dbSNP v.146 (https://www.ncbi.nlm.nih.gov/snp/), (3) variants with conflicting reference alleles (Illumina vs. dbSNP), (4) samples with discordant sex (registry records vs. genotypes), (5) variants with missingness >5%, (6) samples with missingness >2%, (7) variants that failed Hardy–Weinberg test at P < 1 × 10−6, (8) variants with P < 0.01 in tests of differences in missingness between cases and controls, (9) and variants with batch effect (HSCR and hydrocephalus samples were genotyped on the same batch, while the other controls were genotyped previously). These quality control steps were done with PLINK v1.90b3o [20]. We also removed related individuals using KING [21] as implemented in VCFtools [22], up until third-degree relatives, maintaining the samples with lower missingness. European ancestry outliers were removed using the R package SNPRelate v1.2.0 [23]. Outliers were defined as samples that had principal component analysis 1 (PCA1) and/or PCA2 absolute values larger than six times the interquartile range of the mean PCA1 and/or PCA2 values of all our cases and controls when computed together with the known European samples from the Human Genome Diversity Project [24] (Figure S1A and Figure S1B). Of note, the three clusters separated by eigenvector 2 in Figure S1B tag the common European chromosome 8 inversion [25]. The PCA outliers removal procedure was done until no more samples were removed (two rounds sufficed). After the quality control steps, we ended up with 170 cases and 4717 controls genotyped for 861,837 variants with a total genotyping success rate of 99.8%.
Sensitivity analysis
Since the controls in the discovery analysis were comprised of cases of other diseases with existing data from previous GWAS projects in our group, we performed a sensitivity analysis, where GWAS scans were performed leaving out one sub-group at a time from the controls. This had very little influence on estimates and P values and all results presented are therefore based on the full control set.
GWAS replication
We selected 15 variants for the GWAS replication stage using the criteria described in Supplementary Note 1. Briefly, the replication P value criteria reflects the small size of the HSCR case sample, avoidance of batch effects, considerations on minor allele frequency (MAF) and imputation info score, and prior functional knowledge of HSCR disease. The replication cohort from the USA was genotyped on the Illumina HumanOmni2.5M array, while the Finnish cohort was genotyped on the Illumina HumanCoreChip. The USA cohort was subsequently imputed to the Haplotype Reference Consortium panel (version r1.1) [26], while the Finnish cohort was imputed to the 1000 Genomes reference panel [27]. Since the Swedish cohort did not have genome-wide data, genotyping of the 15 selected replication variants was done using PCR-based KASP genotyping chemistry (LGC Genomics). If any of the selected 15 variants could not get a functioning KASP assay, replacements were done with the following criteria: (1) choose variants with Linkage disequilibrium (LD) r2 > 0.9 in 1000 Genomes European population (EUR 1000 Genomes Phase 1 data) as calculated in HaploReg v4.1 [28]; (2) if more than one appeared, choose the one where its MAF in African (AFR), Native American (AMR), and Asian (ASN) populations were the closest to the original SNP; (3) if there still were ties, choose the SNP with the lowest P value in our GWAS. The variants rs17653445, rs2505994, and rs12534941 were substituted with rs72789806, rs2505990, and rs6959065, respectively. Individuals in the Swedish cohort with >10% missing genotypes were excluded; variants with >2% missing data and with deviations from Hardy–Weinberg equilibrium (P < 0.05) were also excluded. GWAS replicated loci were defined as having P < 1 × 10−5 in the discovery cohort, nominally significant associations in all three replication cohorts (P < 0.05) with the same direction of effects, and a combined meta-analysis P < 5 × 10−8.
Replication of conditional analysis
One variant (rs144432435) was selected for replication genotyping, as it was genome-wide significant in the discovery cohort, when conditioned on the lead replicated RET SNP (rs2505994).
Genotype imputation
We performed imputation of unobserved genotypes using phased haplotypes from the Haplotype Reference Consortium panel (version r1.0) [26] using the Positional Burrows–Wheeler Transform algorithm [29] available on the imputation server at the Wellcome Trust Sanger Institute (https://imputation.sanger.ac.uk/). Phasing was also done at the server prior to imputation with SHAPEIT2 v2.r790 [30].
Association analysis
Variants were kept for association if: (1) imputation info score ≥0.9, (2) MAF >1%, and (3) PLINK INFO score <1.2 (INFO values much >1 can indicate strong departure from Hardy–Weinberg equilibrium). GWAS analysis was done using logistic regression on 6,162,479 imputed SNP dosages under an additive genetic model using PLINK v1.90b3o [20]. We carried out an inverse variance-based fixed-effects meta-analysis of the discovery and replication data, as implemented in PLINK v1.90b3o [20]. Estimation of the inflation factor of the discovery stage P values was done with the function estlambda in GenABEL R package [31]. Regional association plots were created with LocusZoom [32].
Genetic correlation
Genetic correlation analysis of our GWAS of Hirschsprung disease vs. GWAS of other diseases/traits was done with the LD score regression method [33, 34] available at LD Hub (http://ldsc.broadinstitute.org/).
Linkage disequilibrium
LD between pairs of variants was calculated in PLINK v1.90b3o [20]. The dosage genotypes were transformed to hard calls prior to LD calculations.
Heritability
Heritability estimation was done with genome-wide complex trait analysis (GCTA) v1.24.7 [35] on the genotyped QC variants. We first estimated the genetic relationship from all the autosomal SNPs, and then did the GCTA-GREML analysis for a case–control study with a specified disease prevalence of 0.000109 [36].
Case–control prediction
To obtain a predictive genetic model, we used Lasso [37] (least absolute shrinkage and selection operator) regression. To avoid overfitting, the optimal Lasso penalization parameter, λ*, was selected by 10-fold cross-validation (details below). Then, based on all variants with a marginal association P < 0.01 (62,652 variants), a predictive genetic model was obtained using Lasso logistic regression with penalization parameter λ*. Finally, the performance of this predictive model was evaluated in an independent validation cohort (USA HSCR cohort). Cross-validation procedure for λ*: Since the variable selection by Lasso is preceded by a P value-based filtering step, it is important that the cross-validation procedure used to select the penalization parameter λ* appropriately reflects this filtering to avoid overfitting. Briefly, the discovery GWAS cohort was first randomly partitioned into 10 equally sized subsamples. A single subsample was labeled as the test data and reserved for quantifying the predictive performance, while a new marginal association analysis was carried out (as described in section “Association analysis” above) on the remaining training data. All variants with a marginal association P > 0.01 were filtered out and a Lasso logistic regression model was fitted to the remaining variants in the training data for a decreasing sequence of penalization parameters; λ1, …, λ200. For each value of λ, the predictive performance of the model was evaluated on the test data in terms of the area under the receiver operator characteristics (ROC) curve (AUC). This procedure was repeated 10 times and subsequently the mean cross-validation AUC was calculated for each value of λ. The optimal Lasso penalization parameter, λ*, was selected as the value of λ achieving the highest mean cross-validation AUC.
Results
Study subjects
In order to identify HSCR susceptibility loci in the Danish population, we did a GWAS with 170 cases, pathology confirmed with aganglionosis, and 4717 controls, all of Danish ancestry (see section “Materials and methods”). Male:female ratio was 3.1 in our cases and 1.1 in the controls. Our data did not permit distinction between S-HSCR and L-HSCR. Of note, information on the length of the aganglionosis was also not available in the replication cohorts.
GWAS confirms known RET and SEMA3 susceptibility variants
All samples from the Danish discovery cohort were genotyped on the Illumina OmniExpressExome chip, with missing genotypes imputed from the Haplotype Reference Consortium panel. Association tests based on 1000G imputed chromosome X did not reveal any variants for follow-up and therefore the paper is focused on autosomal variation. Of note, GWAS analysis adjusting for sex was also done with minimal change in the results (data not shown). We tested for differences in allele dosages between cases and controls at 6,162,479 autosomal SNPs (MAF > 1%, r2 imputation info score ≥0.9; see section “Materials and methods”). Our study subjects showed a genomic inflation factor of 1.03 (Figure S2), suggesting a low population stratification. Moreover, the genomic inflation factor was only 1.0005 when the known RET and SEMA3 signals were removed and we therefore did not correct P values by genomic control. We selected 15 SNPs from different loci for replication (see section “Materials and methods”; Supplementary Note 1), of which four replicated in three independent replication cohorts from USA, Sweden, and Finland (Table 1). Of the four replicated SNPs, three were at the RET locus and another at the SEMA3 locus. We selected more than one SNP for replication genotyping at RET locus as this is the most significant locus associated with HSCR and therefore we did not want to miss any possible new hits. When conditioning on the top replicated RET SNP (rs2505994 hg19.chr10:g.43568887T>C), the other two replicated RET SNPs ceased to be significantly associated with HSCR (rs1271546 hg19.chr10:g.43505098A>G P = 0.04; rs10899836 hg19.chr10:g.44105044G>A P = 0.06), despite their relative low LD (r2 of rs2505994−rs1271546 = 0.18; r2 of rs2505994−rs10899836 = 0.03). The three replicated RET locus SNPs are in LD with rs2505998 hg19.chr10:g.43570925A>G, the previously reported lead RET SNP [12] (r2 of rs2505994−rs2505998 = 0.85; r2 of rs1271546−rs2505998 = 0.18; r2 of rs10899836−rs2505998 = 0.06) (Figure S4). Similarly, our replicated SEMA3 variant is also in LD with rs80227144 hg19.chr7:g.84349842C>A, the previously reported lead SEMA3 SNP [12] (r2 of rs117617821−rs80227144 = 0.97) (Figure S5). In line with previous studies based on Caucasian individuals [9, 12], the NRG1 locus only showed suggestive significance in the discovery cohort (lowest P = 9.83 × 10−6 for rs73250444 hg19.chr8:g.32252694A>C) (Figure S3a) and did not replicate (Table 1). The previously reported lead NRG1 SNP in East Asians [12] was even less significantly associated in our data (rs7005606 hg19.chr8:g.32401501T>G; P = 5.22 × 10−5) (Figure S3b). While our top SNP in NRG1 is in relatively low LD with the reported lead SNP in our discovery cohort (rs73250444–rs7005606; r2 = 0.13), conditional analysis of both SNPs revealed similar effects (rs73250444 P = 0.003 adjusting for rs7005606; rs7005606 P = 0.007 adjusting for rs73250444). Moreover, the previously reported lead NRG1 SNP in East Asians [12] (rs7005606) was less significantly associated in our discovery cohort (P = 5.22 × 10−5) and Finnish replication cohort (P = 0.03), while below nominal significance in our US replication cohort (P = 0.29). After removal of the RET and SEMA3 loci the quantile–quantile plot still indicated an excess of low P values (Figure S6), suggesting that additional loci is associated with the disease.
Conditional analysis identifies a new low-frequency HSCR susceptibility variant
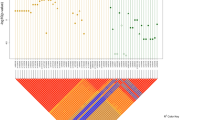
We sought to detect multiple independent association signals at the SEMA3 and RET loci by conditioning on their respective lead SNPs. A secondary association signal was detected at the RET locus (top SNP being rs144432435 hg19.chr10:g.43559390C>T) when we adjusted for the effect of the RET lead SNP rs2505994 (Fig. 1). Of note, this secondary association signal is still of genome-wide significance if we condition on all the three replicated RET SNPs (rs2505994, rs1271546, and rs10899836) (Figure S7). In the Danish discovery cohort, rs144432435 has a frequency of 1% in the controls and 3% in the cases, with a conditional odds ratio (OR) = 6.8, P = 2.7 × 10−8. Before conditional analysis, the effect of rs144432435 was masked, as its risk allele (T) is out of phase with the rs250994 risk allele (T), i.e., they are not on the same haplotype, resulting in an underestimation of the effect sizes of both SNPs if considered individually (Table 2). The conditional analysis was replicated in the Swedish cohort, with a conditional OR = 5.9, P = 8.3 × 10−3. Conditional meta-analysis of the Danish and Swedish cohorts yielded a conditional OR = 6.6, P = 7.7 × 10−10 (Table 2). Since this low-frequency variant was not present in the Finnish cohort and not replicated in the US cohort (P = 0.58), we hypothesize that rs144432435 is tagging a rare haplotype present in Denmark and Sweden but not in the USA. Further studies are needed to determine the presence of the novel HSCR association signal in populations of different origin and experimental studies are also needed to uncover the functional mechanism underlying the association [38]. Similarly, a previous study [12] has also detected an Asian-specific secondary HSCR association signal of low frequency at the RET locus. This provides further evidence that the RET locus may still contain low frequency or rare variants associated with HSCR that remain to be discovered by increasing sample size and/or performing full sequencing of the RET locus.
Regional conditional association plot of the new genome-wide significant HSCR variant. a Association results at RET locus before the conditional analysis. b Association results when conditioning on the RET lead SNP (rs2505994). Color-coded linkage disequilibrium is shown for the top conditional associated SNP rs144432435—in purple). The x-axis represents the chromosome 10 region (hg19 assembly) surrounding the new hit, while the y-axis represents the strength of the association in −log10 (P value) (see color fig online)
Genetic correlation
Since ulcerative colitis has been found to be associated with HSCR [39], we sought to calculate their genetic correlation by using the LD score regression method [33, 34] on our HSCR GWAS vs. a publicly available ulcerative colitis GWAS [39]. We detected a positive genetic correlation of 0.23, although not statistically significant (P = 0.09). Future studies are needed to investigate possible shared genetic architecture of these diseases [38, 40].
Heritability estimation
Using th GCTA tool [35] on the set of genotyped SNPs, we estimated the heritability to be 88% ± standard error (SE) 13% (P = 6.1 × 10−10). This heritability estimate is in line with previous family-based estimates of HSCR heritability [1], indicating that most of the genetic component of the disease is captured by the genotyped variants. This estimate contrasts with the recent heritability estimate of an Asian HSCR GWAS [12] (also using GCTA) of 34% ± SE 11% (P = 3.0 × 10−3). Since the heritability estimate of the Asian GWAS [12] was performed only for variants with MAF >5%, we redid the heritability estimate of our discovery cohort with just MAF >5%, which resulted in 79% ± S.E. 11% (P = 2.6 × 10−13) of variance explained. This heritability estimate is still much higher than what is reported in the Asian GWAS [12], and suggests most of the HSCR heritability in our cohort is tagged by common variation. This discrepancy in heritability estimates between the two populations might indicate (1) different genetic architecture of HSCR in Europeans vs. Asians, (2) SNP chips tagging common variation better in populations of European ancestry [41], and/or (3) different case selection procedures (e.g., length of aganglionosis, male:female ratio, prevalence of HSCR), which all might play a role when calculating heritability estimates.
Case–control prediction
To investigate the possibility of predicting Hirschsprung disease based on a genetic prediction model, we first cross-validated a Lasso prediction approach in the discovery stage data (see section “Materials and methods”). The procedure obtained a mean cross-validation AUC of 81% for the optimal penalization parameter λ* = 0.0114. Based on this penalization, 10 variants were selected by the final Lasso regression on the full discovery stage data. Selected variants and loadings are provided in Table S1. We investigated how the predictive performance of this model generalize in the independent US cohort. Here we obtained a validation AUC of 76% (Fig. 2). The reduced case–control prediction accuracy in the US replication cohort might mean that the Lasso selected variants could be tagging underlying causal variants less well here than in the Danish ancestry discovery cohort. We note that five of the 10 variants are in the RET locus, one is the lead SEMA3 variant, while the remaining four are in loci that also were selected for replication (Table S1). For most of these loci the association signal was attenuated in the US cohort (Table 1).
Regulatory enrichment analysis
We have also mapped the H3K4me1, H3K27ac ChIP-seq peak, and DNase I hypersensitive sites from the human fetal large intestine datasets obtained from the NIH Roadmap Epigenomics Project and the 225 kb topologically associated domain (TAD), which is common across nine cell lines as described previously [42]. Our lead RET SNP (rs2505994) and the newly identified variant (rs144432435) both fall within the TAD, although they do not seem to overlap any of the open chromatin regions or putative enhancer marks (Figure S8).
Discussion
In this study, we confirmed the previously reported RET and SEMA3 loci [9, 10, 12] as being associated with HSCR in our Danish cohort. However, we could not replicate the NRG1 locus previously reported to be associated with HSCR in East Asian populations [11, 12]. Additionally, we detected and replicated a novel low-frequency variant, rs144432435, which was genome-wide significantly associated with HSCR, when conditioning on our RET lead SNP, rs2505994. Furthermore, we estimated the heritability of HSCR, based on the genotyped SNPs, to be 88%, close to classical family study-based estimates [1]. We were able to predict HSCR case–control status with good accuracy based on a subset of 10 SNPs (AUC = 0.76), which might prove useful as an add-on to a first tier HSCR diagnostic [43]. In conclusion, we combined the largest collection of sporadic Hirschsprung cases to date (586 cases) to further elucidate the genetic architecture of this rare disease.
The GWAS summary statistics from our study are available at http://danishnationalbiobank.com/gwas.html.
References
Badner JA, Sieber WK, Garver KL, Chakravarti A. A genetic study of Hirschsprung disease. Am J Hum Genet. 1990;46:568–80.
Löf Granström A, Svenningsson A, Hagel E, Oddsberg J, Nordenskjöld A, Wester T. Maternal risk factors and perinatal characteristics for Hirschsprung disease. Pediatrics. 2016;138:e20154608.
Heanue TA, Pachnis V. Enteric nervous system development and Hirschsprung’s disease: advances in genetic and stem cell studies. Nat Rev Neurosci. 2007;8:466–79.
Parisi MA, Kapur RP. Genetics of Hirschsprung disease. Curr Opin Pediatr. 2000;12:610–7.
Burkardt DD, Graham JM Jr, Short SS, Frykman PK. Advances in Hirschsprung disease genetics and treatment strategies: an update for the primary care pediatrician. Clin Pediatr. 2014;53:71–81.
Amiel J, Sproat-Emison E, Garcia-Barcelo M, et al. Hirschsprung disease, associated syndromes and genetics: a review. J Med Genet. 2008;45:1–14.
Sergi C. Hirschsprung’s disease: historical notes and pathological diagnosis on the occasion of the 100(th) anniversary of Dr. Harald Hirschsprung’s death. World J Clin Pediatr. 2015;4:120–5.
Alves MM, Sribudiani Y, Brouwer RW, et al. Contribution of rare and common variants determine complex diseases-Hirschsprung disease as a model. Dev Biol. 2013;382:320–9.
Jiang Q, Arnold S, Heanue T, et al. Functional loss of semaphorin 3C and/or semaphorin 3D and their epistatic interaction with ret are critical to Hirschsprung disease liability. Am J Hum Genet. 2015;96:581–96.
Emison ES, McCallion AS, Kashuk CS, et al. A common sex-dependent mutation in a RET enhancer underlies Hirschsprung disease risk. Nature. 2005;434:857–63.
Garcia-Barcelo MM, Tang CS, Ngan ES, et al. Genome-wide association study identifies NRG1 as a susceptibility locus for Hirschsprung’s disease. Proc Natl Acad Sci USA. 2009;106:2694–9.
Tang CS, Gui H, Kapoor A, et al. Trans-ethnic meta-analysis of genome-wide association studies for Hirschsprung disease. Hum Mol Genet. 2016;25:5265–75.
Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–53.
Bjerregaard B, Larsen OB. The Danish Pathology Register. Scand J Public Health. 2011;39:72–4.
Pedersen CB. The Danish Civil Registration System. Scand J Public Health. 2011;39:22–5.
Feenstra B, Pasternak B, Geller F, et al. Common variants associated with general and MMR vaccine-related febrile seizures. Nat Genet. 2014;46:1274–82.
Geller F, Feenstra B, Carstensen L, et al. Genome-wide association analyses identify variants in developmental genes associated with hypospadias. Nat Genet. 2014;46:957–63.
Nørgaard-Pedersen B, Hougaard DM. Storage policies and use of the Danish Newborn Screening Biobank. J Inherit Metab Dis. 2007;30:530–6.
Hollegaard MV, Grauholm J, Børglum A, et al. Genome-wide scans using archived neonatal dried blood spot samples. BMC Genom. 2009;10:297.
Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7.
Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–73.
Danecek P, Auton A, Abecasis G, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–8.
Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics. 2012;28:3326–8.
Jakobsson M, Scholz SW, Scheet P, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003.
Ma J, Amos CI. Investigation of inversion polymorphisms in the human genome using principal components analysis. PLoS ONE. 2012;7:e40224.
McCarthy S, Das S, Kretzschmar W, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48:1279–83.
1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526:68–74.
Ward LD, Kellis M. HaploRegv4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44:D877–81.
Durbin R. Efficient haplotype matching and storage using the positional Burrows–Wheeler transform (PBWT). Bioinformatics. 2014;30:1266–72.
Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10:5–6.
Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–6.
Pruim RJ, Welch RP, Sanna S, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7.
Bulik-Sullivan BK, Loh PR, Finucane HK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–5.
Bulik-Sullivan B, Finucane HK, Anttila V, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–41.
Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82.
Best KE, Addor MC, Arriola L, et al. Hirschsprung’s disease prevalence in Europe: a register based study. Birth Defects Res A. 2014;100:695–702.
Tibshirani R. Regression shrinkage and selection via the Lasso. J R Stat Soc B. 1996;58:267–88.
Armstrong GR, Raafat F. Humoral reaction in the inflamed colon in Hirschsprung’s disease and ulcerative colitis. J Clin Pathol. 1988;41:975–7.
Liu JZ, van Sommeren S, Huang H, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–86.
Cucino C, Sonnenberg A. The comorbid occurrence of other diagnoses in patients with ulcerative colitis and Crohn’s disease. Am J Gastroenterol. 2001;96:2107–12.
Ha NT, Freytag S, Bickeboeller H. Coverage and efficiency in current SNP chips. Eur J Hum Genet. 2014;22:1124–30.
Chatterjee S, Kapoor A, Akiyama JA, et al. Enhancer variants synergistically drive dysfunction of a gene regulatory network in Hirschsprung disease. Cell. 2016;167:355–68.
Hackam DJ, Reblock KK, Redlinger RE, Barksdale EM Jr. Diagnosis and outcome of Hirschsprung’s disease: does age really matter? Pediatr Surg Int. 2004;20:319–22.
Acknowledgements
We wish to thank the International Hirschsprung Disease Consortium for sharing their data. This study was funded by the Danish Medical Research council (DFF 4004-00512) and has been conducted using the Danish National Biobank resource. The Danish National Biobank is supported by the Novo Nordisk Foundation. BF was supported by an Oak Foundation fellowship. AC was supported by NIH grant HD28088.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Fadista, J., Lund, M., Skotte, L. et al. Genome-wide association study of Hirschsprung disease detects a novel low-frequency variant at the RET locus. Eur J Hum Genet 26, 561–569 (2018). https://doi.org/10.1038/s41431-017-0053-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-017-0053-7
This article is cited by
-
Hirschsprung disease
Nature Reviews Disease Primers (2023)
-
Genetics of Hirschsprung’s disease
Pediatric Surgery International (2023)
-
Whole genome sequencing reveals epistasis effects within RET for Hirschsprung disease
Scientific Reports (2022)
-
Stem cell-based therapy for hirschsprung disease, do we have the guts to treat?
Gene Therapy (2022)
-
Increased miR-214 expression suppresses cell migration and proliferation in Hirschsprung disease by interacting with PLAGL2
Pediatric Research (2019)