Abstract
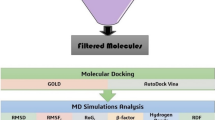
In pursuit of novel antibiotics that could curb the growing trend of multidrug resistance by Salmonella typhimurium, a data set of some cephalosporin analogues were subjected to Molecular Docking based virtual screening against a penicillin-binding protein (PBP 1b) of the bacterium to ascertain the binding affinity values of the bioactive ligands against the active sites of the PBP 1b protein target using the AutoDock Vina Software. Three compounds with binding affinity values ranging from −7.8 kcal/mol to −8.2 kcal/mol were selected as the most promising leads. The selected compounds also displayed better potencies against the bacterium when compared with Cefuroxime (binding affinity = −6.4 kcal/mol), a standard β-lactam antibiotic used herein for quality control and assurance. Furthermore, evaluation of the drug-likeness and ADMET properties of the three most promising leads revealed that they possess good oral bioavailability and excellent pharmacokinetic profiles. It is hoped that the findings of this study will provide an excellent template for developing more potent β-lactam antibiotics against Salmonella typhimurium.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
Change history
24 November 2023
This article has been retracted. Please see the Retraction Notice for more detail: https://doi.org/10.1038/s41429-023-00684-1
References
Vlad MI, Nuta CD, Cornel, Miron CC, et al. In silico and in vitro experimental studies of new Dibenz[b,e]oxepin-11(6H)one O-(arylcarbamoyl)-oximes designed as potential antimicrobial agents. Molecules. 2020, 25; https://doi.org/10.3390/molecules25020321
De Kraker ME, Stewardson AJ, Harbarth S. Will 10 million people die a year due to antimicrobial resistance by 2050? PLoS Med 2016;13:e1002184.
O’Neill J. Review on Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations; Review on Antimicrobial Resistance: London, UK, 2014.
Scherer CA, Miller SI. Molecular pathogenesis of salmonellae. In: Groisman EA (ed.) Principles of bacterial pathogenesis. Academic, New York, 2001, 266-33.
Grein T, O’Flanagan D, McCarthy T, Bauer D. An outbreak of multidrug-resistant Salmonella typhimurium food poisoning at a wedding reception. Ir Med J 1999;92:238–41.
Helms M, Ethelberg S, Molbak K. International Salmonella Typhimurium DT104 infections, 1992–2001. Emerg Infect Dis. 2005;11:859–67.
Crump JA, Medalla FM, Joyce KW, Krueger AL, Hoekstra RM, Whichard JM, Barzilay EJ. Antimicrobial resistance among invasive nontyphoidal Salmonella enterica isolates in the United States: national antimicrobial resistance monitoring system, 1996 to 2007. Antimicrob Agents Chemother 2011;55:1148–54.
Meakins S, Fisher IS, Berghold C, et al. Antimicrobial drug resistance in human nontyphoidal Salmonella isolates in Europe 2000–2004: a report from the Enter-net International Surveillance Network. Microb Drug Resist. 2008;14:31–35.
Hohmann EL. Nontyphoidal salmonellosis. Clin Infect Dis. 2001;15:263–9.
Saha RN, Raman M. Modeling of biological activity and pharmacokinetics of cefixime. Indian J Pharm Edu Res. 2005;42:207–14.
Harrison CJ, Bratcher D. Cephalosporins: A review. Off J Acad Ped. 2008;29:264–73.
Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev 2008;32:234–58.
Navarre WW, Schneewind O. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol Mol Biol Rev. 1999;63:174–229. https://doi.org/10.1128/MMBR.63.1.174-229.1999
Banzhaf M, Van den Berg van Saparoea B, Terrak M, et al. Cooperativity of pepdidoglycan synthases active in bacteria cell elongation. Mol Microbiol. 2012;85:179–94. https://doi.org/10.1111/j.1365-2958.2012.08103.x
Bertsche U, Breukink E, Kast T, Wolf B, et al. Interaction between two murein (pepdidoglycan) synthases PBP3 and PBP1B, in E.coli. Mol Microbiol. 2006;61:675–90. https://doi.org/10.1111/j.1365-2958.2006.05280
Mueller AE, Egan JFA, Breukink E, Vollmer W, Levin AP. Plasticity of E. coli cell wall metabolism promotes fitness and antibiotic resistance across environmental conditions. eLife. 2019;8:e40754. https://doi.org/10.7554/eLife.40754
Shapiro S, Guggenheim B. Inhibition of oral bacteria by phenolic compounds. Part 1. QSAR analysis using molecular connectivity. Quant Struct -Act Relat 1998;17:327–37.
Kore PP, Mutha MM, Antre VR, Oswal JR, Kshirsagar SS. Computer-aided drug design: an innovative tool for modeling. Open J Med Chem. 2012;2:139–48. https://doi.org/10.4236/ojmc.2012.24017
Padole SS, Asnani JA, Chaple DR, Katre GS. A review of approaches in computer-aided drug design in drug discovery. GSC Biol Pharm Sci. 2022;19:075–83. https://doi.org/10.30574/gscbps.2022.19.2.0161
Hossain S, Sarkar B, Prottoy MNI, Araf Y, Taniya MA, Ullah MA. Thrombolytic activity, drug-likeness property and ADME/T analysis of isolated phytochemicals from ginger (Zingiber officinale) using in silico approaches. Mod Res Inflamm. 2019;8:29–43.
Yu H, Adedoyin A. ADME–Tox in drug discovery: Integration of experimental and computational technologies. Drug Discov Today. 2003;8:852–61.
Abishad P, Niveditha P, Unni V, Vergis J, Kurkure NV, Chaudhari S, Rawool DB, Barbuddhe SB. In silico molecular docking and in vitro antimicrobial efficacy of phytochemicals against multi‑drug‑resistant enteroaggregative Escherichia coli and non‑typhoidal Salmonella spp. Gut Pathog. 2021;13:46. https://doi.org/10.1186/s13099-021-00443-3
Almeida FA, Pinto UM, Vanetti MCD. Novel insights from molecular docking of SdiA from Salmonella Enteritidis and Escherichia coli with quorum sensing and quorum quenching molecules. Microb Pathog. 2016;99:178e190.
Durhan B, Yalçın E, Çavuşoğlu K, Acar A. Molecular docking assisted biological functions and phytochemical screening of Amaranthus lividus L. extract. Sci Rep. 2022;12:4308. https://doi.org/10.1038/s41598-022-08421-8
Nagasinduja V, Shahitha S, Prakash B, Kumar DJ. Molecular docking analysis of beta-lactamase from Salmonella species with eicosane. Bioinformation. 2022;18:669–74.
Ameji JP, Uzairu U, Shallangwa GA, Uba S Virtual screening of novel pyridine derivatives as effective inhibitors of DNA gyrase (GyrA) of Salmonella typhi, Curr Chem Lett., 2022, 12, 1–16.
Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–61. https://doi.org/10.1002/jcc.21334
Lipinski CA. Lead and drug-likeness compounds: the rule of five revolution. Drug Discov Today Technol. 2004;1:337–41. https://doi.org/10.1016/j.ddtec.2004
Jasmine SKMD, Reddy VSG, Gorityala N, Sagurthi SR, Mungapati S, Manikanta KN, Allam US. In silico modeling and docking analysis of CTX-M-5, Cefotaxime-Hydrolyzing β Lactamase from human-associated Salmonella Typhimurium. J Pharmacol Pharmacother. 2022;13:135–47.
Konig F, Muller F. Transporters and drug-drug interactions: Important determinants of drug disposition and effects. Phamacol Rev. 2013;65:944–66.
Slaughter RL, Edwards DJ. Recent advances: the cytochrome P450 enzymes. Ann Pharmacother. 1995;29:619–24.
Acknowledgements
We appreciate the technical support of the staff and Postgraduate students of the Physical Chemistry Unit of Ahmadu Bello University Zaria, Nigeria.
Author information
Authors and Affiliations
Contributions
The research was designed by AU, and supervised by AGS and SU. JPA carried out the computational analysis and drafted the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article has been retracted. Please see the retraction notice for more detail: https://doi.org/10.1038/s41429-023-00684-1
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ameji, P.J., Uzairu, A., Shallangwa, G.A. et al. RETRACTED ARTICLE: Molecular docking simulation, drug-likeness assessment, and pharmacokinetic study of some cephalosporin analogues against a penicillin-binding protein of Salmonella typhimurium. J Antibiot 76, 211–224 (2023). https://doi.org/10.1038/s41429-023-00598-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41429-023-00598-y