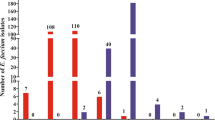
Abstract
Eravacycline (Erava) is a synthetic fluorocycline with potent antimicrobial activity against a wide range of Gram-positive bacteria. This study aimed to investigate the in vitro antimicrobial activity and resistance mechanism of Erava in clinical E. faecium isolates from China. Erava minimum inhibitory concentrations (MICs) against clinical E. faecium isolates—including those resistant to linezolid (LZD) or harboring the tetracycline (Tet) resistance genes was ≤0.25 mg l−1. Moreover, our data indicated that clinical isolates of E. faecium with Erava MIC 0.25 mg l−1 were predominantly shown to belong to Sequence-type 78 (ST78) and ST80. The prevalence of Erava heteroresistance in clinical E. faecium strain was 2.46% (3/122). The increased Erava MIC values of heteroresistance-derived E. faecium clones could be significantly reduced by efflux pump inhibitors (EPIs). Furthermore, comparative proteomics results showed that efflux pumps lmrA, mdlA, and mdlB contributed significantly to the acquisition of Erava resistance in E. faecium. In addition, a genetic mutation in 16 S rRNA (G190A) were detected in resistant E. faecium isolates induced by Erava. In summary, Erava exhibits potent in vitro antimicrobial activity against E. faecium, but mutation of Tet target sites and elevated expression of efflux pumps under Erava selection results in Erava resistance.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Fiore E, Van Tyne D, Gilmore MS. Pathogenicity of Enterococci. Microbiol Spectr. 2019;7:GPP3-0053-2018.
Gao W, Howden BP, Stinear TP. Evolution of virulence in Enterococcus faecium, a hospital-adapted opportunistic pathogen. Curr Opin Microbiol. 2018;41:76–82.
Arias CA, Murray BE. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol. 2012;10:266–78.
Lebreton F, van Schaik W, Manson McGuire A, Godfrey P, Griggs A, Mazumdar V, Corander J, Cheng L, Saif S, Young S et al: Emergence of Epidemic Multidrug-Resistant Enterococcus faecium from Animal and Commensal Strains. mBio. 2013;4:e00534–00513.
Murray BE. Vancomycin-resistant Enterococcal infections. N Engl J Med. 2000;342:710–21.
Alosaimy S, Abdul-Mutakabbir JC, Kebriaei R, Jorgensen SCJ, Rybak MJ. Evaluation of Eravacycline: a novel fluorocycline. Pharmacotherapy. 2020;40:221–38.
Lee YR, Burton CE. Eravacycline, a newly approved fluorocycline. Eur J Clin Microbiol Infect Dis. 2019;38:1787–94.
Scott LJ. Eravacycline: a review in complicated intra-abdominal infections. Drugs. 2019;79:315–24.
Thabit AK, Monogue ML, Newman JV, Nicolau DP. Assessment of in vivo efficacy of eravacycline against Enterobacteriaceae exhibiting various resistance mechanisms: a dose-ranging study and pharmacokinetic/pharmacodynamic analysis. Int J Antimicrob Agents. 2018;51:727–32.
Livermore DM, Mushtaq S, Warner M, Woodford N. In vitro activity of eravacycline against carbapenem-resistant Enterobacteriaceae and Acinetobacter baumannii. Antimicrobial agents Chemother. 2016;60:3840–44.
Monogue ML, Thabit AK, Hamada Y, Nicolau DP. Antibacterial efficacy of Eravacycline in vivo against gram-positive and gram-negative organisms. Antimicrobial agents Chemother. 2016;60:5001–5.
Sutcliffe JA, O’Brien W, Fyfe C, Grossman TH. Antibacterial activity of eravacycline (TP-434), a novel fluorocycline, against hospital and community pathogens. Antimicrobial agents Chemother. 2013;57:5548–58.
Zhang F, Bai B, Xu GJ, Lin ZW, Li GQ, Chen Z, Cheng H, Sun X, Wang HY, Chen, Zheng JX, Deng QW, Yu ZJYW. Eravacycline activity against clinical S. aureus isolates from China: in vitro activity, MLST profiles and heteroresistance. BMC Microbiol. 2018;18:211.
Zheng J, Lin Z, Sun X, Lin W, Chen Z, Wu Y, Qi G, Deng Q, Qu D, Yu Z. Overexpression of OqxAB and MacAB efflux pumps contributes to eravacycline resistance and heteroresistance in clinical isolates of Klebsiella pneumoniae. Emerg Microbes Infect. 2018;7:139.
Nguyen F, Starosta AL, Arenz S, Sohmen D, Donhofer A, Wilson DN. Tetracycline antibiotics and resistance mechanisms. Biol Chem. 2014;395:559–75.
Wen Z, Shang Y, Xu G, Pu Z, Lin Z, Bai B, Chen Z, Zheng J, Deng Q, Yu Z. Mechanism of Eravacycline resistance in clinical Enterococcus faecalis Isolates from China. Front Microbiol. 2020;11:916.
Chen Y, Hu D, Zhang Q, Liao XP, Liu YH, Sun J. Efflux pump overexpression contributes to tigecycline heteroresistance in Salmonella enterica serovar Typhimurium. Front Cell Infect Microbiol. 2017;7:37.
Liu H, Jia X, Zou H, Sun S, Li S, Wang Y, Xia Y. Detection and characterization of tigecycline heteroresistance in E. cloacae: clinical and microbiological findings. Emerg Microbes Infect. 2019;8:564–74.
Caglan E, Nigiz S, Sancak B, Gur D. Resistance and heteroresistance to colistin among clinical isolates of Acinetobacter baumannii. Acta microbiologica et immunologica Hungarica. 2019;52:1–5.
Fiedler S, Bender JK, Klare I, Halbedel S, Grohmann E, Szewzyk U, Werner G. Tigecycline resistance in clinical isolates of Enterococcus faecium is mediated by an upregulation of plasmid-encoded tetracycline determinants tet(L) and tet(M). J antimicrobial Chemother. 2016;71:871–81.
Grossman TH. Tetracycline antibiotics and resistance. Cold Spring Harb Perspect Med. 2016;6:a025387.
Muthaiyan A, Silverman JA, Jayaswal RK, Wilkinson BJ. Transcriptional profiling reveals that daptomycin induces the Staphylococcus aureus cell wall stress stimulon and genes responsive to membrane depolarization. Antimicrobial agents Chemother. 2008;52:980–90.
Reyes J, Panesso D, Tran TT, Mishra NN, Cruz MR, Munita JM, Singh KV, Yeaman MR, Murray BE, Shamoo, Garsin D, Bayer AS, Arias CAY. A liaR deletion restores susceptibility to daptomycin and antimicrobial peptides in multidrug-resistant Enterococcus faecalis. J Infect Dis. 2015;211:1317–25.
Scherl A, François P, Charbonnier Y, Deshusses JM, Koessler T, Huyghe A, Bento M, Stahl-Zeng J, Fischer A, Masselot, Vaezzadeh A, Gallé F, Renzoni A, Vaudaux P, Lew D, Zimmermann-Ivol CG, Binz PA, Sanchez JC, Hochstrasser DF, Schrenzel JA. Exploring glycopeptide-resistance in Staphylococcus aureus: a combined proteomics and transcriptomics approach for the identification of resistance-related markers. BMC genomics. 2006;7:296.
Drummelsmith J, Winstall E, Bergeron MG, Poirier GG, Ouellette M. Comparative proteomics analyses reveal a potential biomarker for the detection of vancomycin-intermediate Staphylococcus aureus strains. J proteome Res. 2007;6:4690–702.
Jousselin A, Renzoni A, Andrey DO, Monod A, Lew DP, Kelley WL. The posttranslocational chaperone lipoprotein PrsA is involved in both glycopeptide and oxacillin resistance in Staphylococcus aureus. Antimicrobial agents Chemother. 2012;56:3629–40.
Zheng J-X, Lin Z-W, Chen C, Chen Z, Lin F-J, Wu Y, Yang S-Y, Sun X, Yao W-M, Li D-Y, Yu Z-J, Jin J-L, Qu D, Deng Q-W. Biofilm formation in Klebsiella pneumoniae bacteremia strains was found to be associated with CC23 and the presence of wcaG. Front Cell Infect Microbiol. 2018;8:21.
Institute CaLS: Performance standards for antimicrobial susceptibility testing: 24th Informational supplement. Document M100-S26. Wayne, PA: Clinical and Laboratory Standards Institute 2016.
Bai B, Lin Z, Pu Z, Xu G, Zhang F, Chen Z, Sun X, Zheng J, Li P, Deng, Yu ZQ. In vitro activity and heteroresistance of omadacycline against clinical Staphylococcus aureus isolates from China reveal the impact of Omadacycline susceptibility by branched-chain amino acid transport system II carrier protein, Na/Pi cotransporter family protein, and fibronectin-binding protein. Front Microbiol. 2019;10:2546.
Bai B, Hu K, Li H, Yao W, Li D, Chen Z, Cheng H, Zheng J, Pan W, Deng M, Liu X, Lin Z, Deng Q, Yu Z. Effect of tedizolid on clinical Enterococcus isolates: in vitro activity, distribution of virulence factor, resistance genes and multilocus sequence typing. FEMS Microbiol Lett. 2018;365:fnx284.
Linkevicius M, Sandegren L, Andersson DI. Potential of tetracycline resistance proteins to evolve tigecycline resistance. Antimicrobial agents Chemother. 2016;60:789–96.
Sader HS, Castanheira M, Farrell DJ, Flamm RK, Mendes RE, Jones RN. Tigecycline antimicrobial activity tested against clinical bacteria from Latin American medical centres: results from SENTRY Antimicrobial Surveillance Program (2011-2014). Int J Antimicrob Agents. 2016;48:144–50.
Ahmed MO, Baptiste KE. Vancomycin-resistant enterococci: a review of antimicrobial resistance mechanisms and perspectives of human and animal health. Microb drug resistance (Larchmt, NY). 2018;24:590–606.
Bassetti M, Corey R, Doi Y, Morrissey I, Grossman T, Olesky M, Scoble P, Sutcliffe J. In Vitro Global Surveillance of Eravacycline and Comparators Against Staphylococcus spp. and Enterococcus spp. Over a 3-Year Period (2013–2015). Open Forum Infectious Diseases. 2016;3.
Lagace-Wiens PRS, Adam HJ, Laing NM, Baxter MR, Martin I, Mulvey MR, Karlowsky JA, Hoban DJ, Zhanel GG. Antimicrobial susceptibility of clinical isolates of Neisseria gonorrhoeae to alternative antimicrobials with therapeutic potential. J antimicrobial Chemother. 2017;72:2273–7.
Solomkin JS, Ramesh MK, Cesnauskas G, Novikovs N, Stefanova P, Sutcliffe JA, Walpole SM, Horn PT. Phase 2, randomized, double-blind study of the efficacy and safety of two dose regimens of eravacycline versus ertapenem for adult community-acquired complicated intra-abdominal infections. Antimicrobial agents Chemother. 2014;58:1847–54.
Zhanel GG, Baxter MR, Adam HJ, Sutcliffe J, Karlowsky JA. In vitro activity of eravacycline against 2213 Gram-negative and 2424 Gram-positive bacterial pathogens isolated in Canadian hospital laboratories: CANWARD surveillance study 2014-2015. Diagn Microbiol Infect Dis. 2018;91:55–62.
Lebreton F, Manson AL, Saavedra JT, Straub TJ, Earl AM, Gilmore MS. Tracing the Enterococci from paleozoic origins to the hospital. Cell. 2017;169:849–61.e813.
Freitas AR, Novais C, Ruiz-Garbajosa P, Coque TM, Peixe L. Dispersion of multidrug-resistant Enterococcus faecium isolates belonging to major clonal complexes in different Portuguese settings. Appl Environ Microbiol. 2009;75:4904–8.
Ahmed M, Elramalli A, Baptiste K, Daw M, Zorgani A, Brouwer E, Willems R, Top J. Whole Genome sequence analysis of the first vancomycin-resistant enterococcus faecium isolates from a Libyan hospital in Tripoli. Microb Drug Resist. 2020;26:1390–8.
Eisenberger D, Tuschak C, Werner M, Bogdan C, Bollinger T, Hossain H, Friedrich P, Hussein Z, Pöhlmann C, Würstl B, Nickel S, Lehner-Reindl V, Höller C, Liebl B, Valenza G. Whole-genome analysis of vancomycin-resistant Enterococcus faecium causing nosocomial outbreaks suggests the occurrence of few endemic clonal lineages in Bavaria, Germany. J antimicrob Chemother. 2020;75:1398–404.
Khan MA, Northwood JB, Loor RGJ, Tholen ATR, Riera E, Falcón M, Network PA, van Belkum A, van Westreenen M, Hays JP. High prevalence of ST-78 infection-associated vancomycin-resistant Enterococcus faecium from hospitals in Asunción, Paraguay. Clin Microbiol Infect. 2010;16:624–7.
Sun L, Xu J, Wang W, He F. Emergence of vanA-type vancomycin-resistant Enterococcus faecium ST 78 Strain with a rep2-type plasmid carrying a Tn1546-like element isolated from a urinary tract infection in China. Infect drug resistance. 2020;13:949–55.
Beabout K, Hammerstrom TG, Perez AM, Magalhaes BF, Prater AG, Clements TP, Arias CA, Saxer G, Shamoo Y. The ribosomal S10 protein is a general target for decreased tigecycline susceptibility. Antimicrobial agents Chemother. 2015;59:5561–6.
Niebel M, Quick J, Prieto AMG, Hill RLR, Pike R, Huber D, David M, Hornsey M, Wareham D, Oppenheim, Woodford N, van Schaik W, Loman NB. Deletions in a ribosomal protein-coding gene are associated with tigecycline resistance in Enterococcus faecium. Int J Antimicrob Agents. 2015;46:572–5.
Lee CR, Lee JH, Park KS, Jeong BC, Lee SH. Quantitative proteomic view associated with resistance to clinically important antibiotics in Gram-positive bacteria: a systematic review. Front Microbiol. 2015;6:828.
Shi Y, Hua X, Xu Q, Yang Y, Zhang L, He J, Mu X, Hu L, Leptihn S, Yu Y. Mechanism of eravacycline resistance in Acinetobacter baumannii mediated by a deletion mutation in the sensor kinase adeS, leading to elevated expression of the efflux pump AdeABC. Infect Genet Evol. 2020;80:104185.
Xu Y, Tang Y, Wang N, Liu J, Cai X, Cai H, Li J, Tan G, Liu R, Bai, Zhang L, Wu H, Zhang BL. Transcriptional regulation of a leucine-responsive regulatory protein for directly controlling lincomycin biosynthesis in Streptomyces lincolnensis. Appl Microbiol Biotechnol. 2020;104:2575–87.
Pieper R, Gatlin-Bunai CL, Mongodin EF, Parmar PP, Huang ST, Clark DJ, Fleischmann RD, Gill SR, Peterson SN. Comparative proteomic analysis of Staphylococcus aureus strains with differences in resistance to the cell wall-targeting antibiotic vancomycin. Proteomics. 2006;6:4246–58.
Funding
This work was supported by the following grants: National Natural Science Foundation of China (82172283); Natural Science Foundation of Guangdong Province, China (2020A1515011049, 2020A1515111146, 2021A1515011727); Sanming Project of Medicine in Shenzhen (SMGC201705029); Shenzhen Key Medical Discipline Construction Fund (SZXK06162); Science, Technology and Innovation Commission of Shenzhen Municipality of basic research funds (JCYJ20180302144721183, JCYJ20180302144345028, JCYJ20190809110622729, JCYJ20190809110209389, JCYJ20190809102219774, JCYJ20190809151817062) and the Shenzhen Nanshan District Scientific Research Program of the People’s Republic of China (NS009, NS117, NS140, NS144, NS066, 2020012, 2020018, 2020040, 20200598).
Author information
Authors and Affiliations
Contributions
ZC, QD and ZX participated in the design of the study and revised the manuscript carefully; ZW, FL, PZ and YW collected and identified strains, carried out the antibiotic susceptibility test, efflux inhibition assays and analyzed the data; YS, JZ and GL conducted the induction of Erava-resistance test and proteomics; ZY performed the CCs clustering analysis; ZC, QD, ZX and ZW wrote the manuscript incorporating comments from all authors. All the authors have read and agreed upon the submission.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Shenzhen Nanshan people’s Hospital, the 6th Affiliated Hospital of Shenzhen University Health Science Center. For this type of study, formal consent is not required.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
41429_2022_546_MOESM3_ESM.xlsx
Annotation of differentially expressed proteins between E. faecium FM1 and its derivative Erava-induced resistant isolates FM1-E1
Rights and permissions
About this article
Cite this article
Wen, Z., Liu, F., Zhang, P. et al. In vitro activity and adaptation strategies of eravacycline in clinical Enterococcus faecium isolates from China. J Antibiot 75, 498–508 (2022). https://doi.org/10.1038/s41429-022-00546-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41429-022-00546-2