Abstract
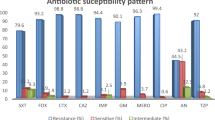
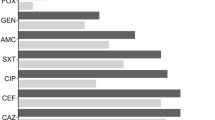
Klebsiella pneumoniae is an opportunistic pathogen that frequently causes nosocomial urinary tract infection (UTI). The aim of this study was to investigate the prevalence of extended-spectrum β-lactamases (ESBL), plasmid-mediated quinolone resistance (PMQR) genes, in acquired AmpC (ac-AmpC) β‑lactamase‑producing K. pneumoniae isolates from patients with nosocomial UTI and to characterize the transmissibility of plasmids harbouring multiple resistance genes. From January 2017 to June 2018, we collected 46 ac-AmpC-producing K. pneumoniae isolates causing UTI from a tertiary care hospital in China. Antimicrobial susceptibility assays showed that non-susceptibility of all isolates to third-generation cephalosporin and fluoroquinolone was very high (>80%). Diverse types of ESBLs and PMQR genes, including blaSHV-12 (n = 23), blaSHV-27 (n = 1), blaSHV-28 (n = 2), blaSHV-33 (n = 4), blaCTX-M-3 (n = 24), blaCTX-M-14 (n = 6), blaCTX-M-15 (n = 6), blaCTX-M-22 (n = 1) and blaOXA-10 (n = 26), as well as qnrA (n = 2), qnrB (n = 39) and qnrS (n = 2) genes were identified amongst AmpC-producing K. pneumoniae isolates. The blaAmpC, qnrB and several ESBLs genes from six strains harbouring multiple AmpC (at least two ampC) were co-transferrable to recipients via conjugation or electroporation, with IncFIA, IncFIB and IncA/C being the dominant replicons. Conserved genetic context associated with the mobilization of blaampC genes was detected. Forty-six isolates were categorized into 25 enterobacterial repetitive intergenic consensus (ERIC) types, and the 6 isolates harbouring multiple AmpC genes belonged to ST1 lineage. This work reports that the emergence of plasmids co-harbouring multiple resistance determinants and mediating the local prevalence in K. pneumoniae causing UTI in China.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol. 2015;13:269–84.
Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998;11:589–603.
Azargun R, Sadeghi MR, Soroush Barhaghi MH, Samadi Kafil H, Yeganeh F, Ahangar Oskouee M, et al. The prevalence of plasmid-mediated quinolone resistance and ESBL-production in Enterobacteriaceae isolated from urinary tract infections. Infect Drug Resist. 2018;11:1007–14.
Livermore DM. beta-Lactamases- the Threat Renews. Curr Protein Pept Sci. 2009;10:397–400.
Ribeiro TG, Novais A, Rodrigues C, Nascimento R, Freitas F, Machado E, et al. Dynamics of clonal and plasmid backgrounds of Enterobacteriaceae producing acquired AmpC in Portuguese clinical settings over time. Int J Antimicrob Agents. 2019;53:650–6.
Maeyama Y, Taniguchi Y, Hayashi W, Ohsaki Y, Osaka S, Koide S, et al. Prevalence of ESBL/AmpC genes and specific clones among the third-generation cephalosporin-resistant Enterobacteriaceae from canine and feline clinical specimens in Japan. Vet Microbiol. 2018;216:183–9.
Dziri O, Dziri R, Maraoub A, Chouchani C. First report of SHV-148-Type ESBL and CMY-42-Type AmpC beta-Lactamase in Klebsiella pneumoniae clinical isolates in Tunisia. Microb Drug Resist. 2018. [Epub ahead of print].
Li Y, Li Q, Du Y, Jiang X, Tang J, Wang J, et al. Prevalence of plasmid-mediated AmpC beta-lactamases in a Chinese university hospital from 2003 to 2005: first report of CMY-2-Type AmpC beta-lactamase resistance in China. J Clin Microbiol. 2008;46:1317–21.
Jacoby GA. AmpC beta-lactamases. Clin Microbiol Rev. 2009;22:161–82. Table of Contents.
Alonso N, Miro E, Pascual V, Rivera A, Simo M, Garcia MC, et al. Molecular characterisation of acquired and overproduced chromosomal blaAmpC in Escherichia coli clinical isolates. Int J Antimicrob Agents. 2016;47:62–8.
Doublet B, Praud K, Nguyen-Ho-Bao T, Argudin MA, Bertrand S, Butaye P, et al. Extended-spectrum beta-lactamase- and AmpC beta-lactamase-producing D-tartrate-positive Salmonella enterica serovar Paratyphi B from broilers and human patients in Belgium, 2008-10. J Antimicrob Chemother. 2014;69:1257–64.
Girlich D, Bonnin RA, Dortet L, Naas T. Genetics of acquired antibiotic resistance genes in proteus spp. Front Microbiol. 2020;11:256.
Denisuik AJ, Lagace-Wiens PR, Pitout JD, Mulvey MR, Simner PJ, Tailor F, et al. Molecular epidemiology of extended-spectrum beta-lactamase-, AmpC beta-lactamase- and carbapenemase-producing Escherichia coli and Klebsiella pneumoniae isolated from Canadian hospitals over a 5 year period: CANWARD 2007-11. J Antimicrob Chemother. 2013;68(Suppl 1):i57–65.
Luk S, Wong WK, Ho AY, Yu KC, To WK, Ng TK. Clinical features and molecular epidemiology of plasmid-mediated DHA-type AmpC beta-lactamase-producing Klebsiella pneumoniae blood culture isolates. Hong Kong J Glob Antimicrob Resist. 2016;7:37–42.
Cheong HS, Chung DR, Lee C, Kim SH, Kang CI, Peck KR, et al. Emergence of serotype K1 Klebsiella pneumoniae ST23 strains co-producing the plasmid-mediated AmpC beta-lactamase DHA-1 and an extended-spectrum beta-lactamase in Korea. Antimicrob Resist Infect Control. 2016;5:50.
Ben Said L, Jouini A, Alonso CA, Klibi N, Dziri R, Boudabous A, et al. Characteristics of extended-spectrum beta-lactamase (ESBL)- and pAmpC beta-lactamase-producing Enterobacteriaceae of water samples in Tunisia. Sci Total Environ. 2016;550:1103–9.
Philippon A, Arlet G, Jacoby GA. Plasmid-determined AmpC-type beta-lactamases. Antimicrob Agents Chemother. 2002;46:1–11.
Meini S, Tascini C, Cei M, Sozio E, Rossolini GM. AmpC beta-lactamase-producing Enterobacterales: what a clinician should know. Infection. 2019;47:363–75.
Ma J, Zeng Z, Chen Z, Xu X, Wang X, Deng Y, et al. High prevalence of plasmid-mediated quinolone resistance determinants qnr, aac(6’)-Ib-cr, and qepA among ceftiofur-resistant Enterobacteriaceae isolates from companion and food-producing animals. Antimicrob Agents Chemother. 2009;53:519–24.
Rodriguez-Martinez JM, Machuca J, Cano ME, Calvo J, Martinez-Martinez L, Pascual A. Plasmid-mediated quinolone resistance: two decades on. Drug Resist Updat. 2016;29:13–29.
Veldman K, Kant A, Dierikx C, van Essen-Zandbergen A, Wit B, Mevius D. Enterobacteriaceae resistant to third-generation cephalosporins and quinolones in fresh culinary herbs imported from Southeast Asia. Int J Food Microbiol. 2014;177:72–7.
Gude MJ, Seral C, Saenz Y, Gonzalez-Dominguez M, Torres C, Castillo FJ. Evaluation of four phenotypic methods to detect plasmid-mediated AmpC beta-lactamases in clinical isolates. Eur J Clin Microbiol Infect Dis. 2012;31:2037–43.
Perez-Perez FJ, Hanson ND. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002;40:2153–62.
CLSI Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Third Informational Supplement. CLSI document M100-S27 (Clinical Laboratory Standards Institute W, PA, USA, (2017).
Xu H, Huo C, Sun Y, Zhou Y, Xiong Y, Zhao Z, et al. Emergence and molecular characterization of multidrug-resistant Klebsiella pneumoniae isolates harboring bla CTX-M-15 extended-spectrum beta-lactamases causing ventilator-associated pneumonia in China. Infect Drug Resist. 2019;12:33–43.
El-Badawy MF, Alrobaian MM, Shohayeb MM, Abdelwahab SF. Investigation of six plasmid-mediated quinolone resistance genes among clinical isolates of pseudomonas: a genotypic study in Saudi Arabia. Infect Drug Resist. 2019;12:915–23.
Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63:219–28.
Turton JF, Perry C, Elgohari S, Hampton CV. PCR characterization and typing of Klebsiella pneumoniae using capsular type-specific, variable number tandem repeat and virulence gene targets. J Med Microbiol. 2010;59:541–7.
Mata C, Miro E, Alvarado A, Garcillan-Barcia MP, Toleman M, Walsh TR, et al. Plasmid typing and genetic context of AmpC beta-lactamases in Enterobacteriaceae lacking inducible chromosomal ampC genes: findings from a Spanish hospital 1999-2007. J Antimicrob Chemother. 2012;67:115–22.
Carattoli A. Plasmids and the spread of resistance. Int J Med Microbiol. 2013;303:298–304.
Donati V, Feltrin F, Hendriksen RS, Svendsen CA, Cordaro G, Garcia-Fernandez A, et al. Extended-spectrum-beta-lactamases, AmpC beta-lactamases and plasmid mediated quinolone resistance in klebsiella spp. from companion animals in Italy. PLoS One. 2014;9:e90564.
Jean SS, Coombs G, Ling T, Balaji V, Rodrigues C, Mikamo H, et al. Epidemiology and antimicrobial susceptibility profiles of pathogens causing urinary tract infections in the Asia-Pacific region: results from the Study for Monitoring Antimicrobial Resistance Trends (SMART), 2010-2013. Int J Antimicrob Agents. 2016;47:328–34.
Li P, Liu D, Zhang X, Tuo H, Lei C, Xie X, et al. Characterization of plasmid-mediated quinolone resistance in gram-negative bacterial strains from animals and humans in China. Micro Drug Resist. 2019;25:1050–6.
Pai H, Seo MR, Choi TY. Association of QnrB determinants and production of extended-spectrum beta-lactamases or plasmid-mediated AmpC beta-lactamases in clinical isolates of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2007;51:366–8.
Naseer U, Haldorsen B, Simonsen GS, Sundsfjord A. Sporadic occurrence of CMY-2-producing multidrug-resistant Escherichia coli of ST-complexes 38 and 448, and ST131 in Norway. Clin Microbiol Infect. 2010;16:171–8.
Wiesner M, Fernandez-Mora M, Cevallos MA, Zavala-Alvarado C, Zaidi MB, Calva E, et al. Conjugative transfer of an IncA/C plasmid-borne blaCMY-2 gene through genetic re-arrangements with an IncX1 plasmid. BMC Microbiol. 2013;13:264.
Carattoli A. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother. 2009;53:2227–38.
Cherif T, Saidani M, Decre D, Boutiba-Ben Boubaker I, Arlet G. Cooccurrence of multiple AmpC beta-Lactamases in Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis in Tunisia. Antimicrob Agents Chemother. 2016;60:44–51.
Verdet C, Benzerara Y, Gautier V, Adam O, Ould-Hocine Z, Arlet G. Emergence of DHA-1-producing Klebsiella spp. in the Parisian region: genetic organization of the ampC and ampR genes originating from Morganella morganii. Antimicrob Agents Chemother. 2006;50:607–17.
Chen F, Zhang W, Schwarz S, Zhu Y, Li R, Hua X, et al. Genetic characterization of an MDR/virulence genomic element carrying two T6SS gene clusters in a clinical Klebsiella pneumoniae isolate of swine origin. J Antimicrob Chemother. 2019;74:1539–44.
Garcia-Fernandez A, Villa L, Moodley A, Hasman H, Miriagou V, Guardabassi L, et al. Multilocus sequence typing of IncN plasmids. J Antimicrob Chemother. 2011;66:1987–91.
Shon AS, Bajwa RP, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013;4:107–18.
Acknowledgements
We thank Dr. Hui Xu for providing PCR controls.
Funding
This work was supported by the National Natural Science Foundation of China (grant number 81401697), the Natural Science Foundation of Liaoning Province (grant number 2019-MS-092), Scientific Research Project of the Education Department of Liaoning Province (grant number LZ2020052) and the Program for High-Level Entrepreneurial and Innovative Talents of Dalian, China (2017RQ120). This work was also supported by the Liaoning Provincial Program for Top Discipline of Basic Medical Sciences.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declares no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Xiong, Y., Zhang, C., Gao, W. et al. Genetic diversity and co-prevalence of ESBLs and PMQR genes among plasmid-mediated AmpC β-lactamase-producing Klebsiella pneumoniae isolates causing urinary tract infection. J Antibiot 74, 397–406 (2021). https://doi.org/10.1038/s41429-021-00413-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41429-021-00413-6