Abstract
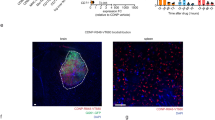
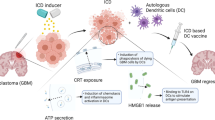
A highly immunosuppressive tumor microenvironment (TME) and the presence of the blood‒brain barrier are the two major obstacles to eliciting an effective immune response in patients with high-grade glioma (HGG). Here, we tried to enhance the local innate immune response in relapsed HGG by intracranially injecting poly(I:C) to establish a robust antitumor immune response in this registered clinical trial (NCT03392545). During the follow-up, 12/27 (44.4%) patients who achieved tumor control concomitant with survival benefit were regarded as responders in our study. We found that the T-cell receptor (TCR) repertoire in the TME was reshaped after poly(I:C) treatment. Based on the RNA-seq analysis of tumor samples, the expression of annexin A1 (ANXA1) was significantly upregulated in the tumor cells of nonresponders, which was further validated at the protein level. In vitro and in vivo experiments showed that ANXA1 could induce the production of M2-like macrophages and microglia via its surface receptor formyl peptide receptor 1 (FPR1) to establish a Treg cell-driven immunosuppressive TME and suppress the antitumor immune response facilitated by poly(I:C). The ANXA1/FPR1 signaling axis can inhibit the innate immune response of glioma patients by promoting an anti-inflammatory and Treg-driven TME. Moreover, ANXA1 could serve as a reliable predictor of response to poly(I:C), with a notable predictive accuracy rate of 92.3%. In light of these notable findings, this study unveils a new perspective of immunotherapy for gliomas.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Ostrom QT, Price M, Neff C, Cioffi G, Waite KA, Kruchko C, et al. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2015-2019. Neuro Oncol. 2022;24:v1–v95. https://doi.org/10.1093/neuonc/noac202.
Tan AC, Ashley DM, Lopez GY, Malinzak M, Friedman HS, Khasraw M. Management of glioblastoma: State of the art and future directions. CA Cancer J Clin. 2020;70:299–312. https://doi.org/10.3322/caac.21613.
Wefel JS, Noll KR, Scheurer ME. Neurocognitive functioning and genetic variation in patients with primary brain tumours. Lancet Oncol. 2016;17:e97–e108. https://doi.org/10.1016/S1470-2045(15)00380-0.
Weyer-Jamora C, Brie MS, Luks TL, Smith EM, Hervey-Jumper SL, Taylor JW. Postacute cognitive rehabilitation for adult brain tumor patients. Neurosurgery. 2021;89:945–53. https://doi.org/10.1093/neuros/nyaa552.
Jackson CM, Choi J, Lim M. Mechanisms of immunotherapy resistance: lessons from glioblastoma. Nat Immunol. 2019;20:1100–9. https://doi.org/10.1038/s41590-019-0433-y.
Arvanitis CD, Ferraro GB, Jain RK. The blood-brain barrier and blood-tumour barrier in brain tumours and metastases. Nat Rev Cancer. 2020;20:26–41. https://doi.org/10.1038/s41568-019-0205-x.
Patel MA, Pardoll DM. Concepts of immunotherapy for glioma. J Neurooncol. 2015;123:323–30. https://doi.org/10.1007/s11060-015-1810-5.
Watowich, MB, Gilbert, MR, and Larion, M. T cell exhaustion in malignant gliomas. Trends Cancer. https://doi.org/10.1016/j.trecan.2022.12.008. (2023).
Kreatsoulas D, Bolyard C, Wu BX, Cam H, Giglio P, Li Z. Translational landscape of glioblastoma immunotherapy for physicians: guiding clinical practice with basic scientific evidence. J Hematol Oncol. 2022;15:80 https://doi.org/10.1186/s13045-022-01298-0.
Jiang H, Yu K, Cui Y, Ren X, Li M, Yang C, et al. Combination of immunotherapy and radiotherapy for recurrent malignant gliomas: Results From a prospective study. Front Immunol. 2021;12:632547 https://doi.org/10.3389/fimmu.2021.632547.
Deng S, Zhu S, Qiao Y, Liu YJ, Chen W, Zhao G, et al. Recent advances in the role of toll-like receptors and TLR agonists in immunotherapy for human glioma. Protein Cell. 2014;5:899–911. https://doi.org/10.1007/s13238-014-0112-6.
De Waele J, Marcq E, Van Audenaerde JR, Van Loenhout J, Deben C, Zwaenepoel K, et al. Poly(I:C) primes primary human glioblastoma cells for an immune response invigorated by PD-L1 blockade. Oncoimmunology. 2018;7:e1407899 https://doi.org/10.1080/2162402X.2017.1407899.
Salmon H, Idoyaga J, Rahman A, Leboeuf M, Remark R, Jordan S, et al. Expansion and activation of CD103(+) dendritic cell progenitors at the tumor site enhances tumor responses to therapeutic PD-L1 and BRAF inhibition. Immunity. 2016;44:924–38. https://doi.org/10.1016/j.immuni.2016.03.012.
Kees T, Lohr J, Noack J, Mora R, Gdynia G, Todt G, et al. Microglia isolated from patients with glioma gain antitumor activities on poly (I:C) stimulation. Neuro Oncol. 2012;14:64–78. https://doi.org/10.1093/neuonc/nor182.
Shime H, Matsumoto M, Oshiumi H, Tanaka S, Nakane A, Iwakura Y, et al. Toll-like receptor 3 signaling converts tumor-supporting myeloid cells to tumoricidal effectors. Proc Natl Acad Sci USA. 2012;109:2066–71. https://doi.org/10.1073/pnas.1113099109.
Wertel I, Surowka J, Polak G, Barczynski B, Bednarek W, Jakubowicz-Gil J, et al. Macrophage-derived chemokine CCL22 and regulatory T cells in ovarian cancer patients. Tumour Biol. 2015;36:4811–7. https://doi.org/10.1007/s13277-015-3133-8.
Rameshbabu, S, Labadie, BW, Argulian, A, and Patnaik, A. Targeting innate immunity in cancer therapy. Vaccines (Basel) 9. https://doi.org/10.3390/vaccines9020138. (2021).
Foo SL, Yap G, Cui J, Lim LHK. Annexin-A1 - A blessing or a curse in cancer? Trends Mol Med. 2019;25:315–27. https://doi.org/10.1016/j.molmed.2019.02.004.
Garcia Pedrero JM, Fernandez MP, Morgan RO, Herrero Zapatero A, Gonzalez MV, Suarez Nieto C, et al. Annexin A1 down-regulation in head and neck cancer is associated with epithelial differentiation status. Am J Pathol. 2004;164:73–9. https://doi.org/10.1016/S0002-9440(10)63098-2.
Rodrigo JP, Garcia-Pedrero JM, Fernandez MP, Morgan RO, Suarez C, Herrero A. Annexin A1 expression in nasopharyngeal carcinoma correlates with squamous differentiation. Am J Rhinol. 2005;19:483–7.
Lin Y, Lin G, Fang W, Zhu H, Chu K. Increased expression of annexin A1 predicts poor prognosis in human hepatocellular carcinoma and enhances cell malignant phenotype. Med Oncol. 2014;31:327. https://doi.org/10.1007/s12032-014-0327-7.
Bai XF, Ni XG, Zhao P, Liu SM, Wang HX, Guo B, et al. Overexpression of annexin 1 in pancreatic cancer and its clinical significance. World J Gastroenterol. 2004;10:1466–70. https://doi.org/10.3748/wjg.v10.i10.1466.
Cheng TY, Wu MS, Lin JT, Lin MT, Shun CT, Huang HY, et al. Annexin A1 is associated with gastric cancer survival and promotes gastric cancer cell invasiveness through the formyl peptide receptor/extracellular signal-regulated kinase/integrin beta-1-binding protein 1 pathway. Cancer. 2012;118:5757–67. https://doi.org/10.1002/cncr.27565.
Schittenhelm J, Trautmann K, Tabatabai G, Hermann C, Meyermann R, Beschorner R. Comparative analysis of annexin-1 in neuroepithelial tumors shows altered expression with the grade of malignancy but is not associated with survival. Mod Pathol. 2009;22:1600–11. https://doi.org/10.1038/modpathol.2009.132.
Yang Y, Liu Y, Yao X, Ping Y, Jiang T, Liu Q, et al. Annexin 1 released by necrotic human glioblastoma cells stimulates tumor cell growth through the formyl peptide receptor 1. Am J Pathol. 2011;179:1504–12. https://doi.org/10.1016/j.ajpath.2011.05.059.
Cheng SX, Tu Y, Zhang S. FoxM1 promotes glioma cells progression by up-regulating Anxa1 expression. PLoS One. 2013;8:e72376. https://doi.org/10.1371/journal.pone.0072376.
Gavins FN, Hickey MJ. Annexin A1 and the regulation of innate and adaptive immunity. Front Immunol. 2012;3:354. https://doi.org/10.3389/fimmu.2012.00354.
Perretti M, D’Acquisto F. Annexin A1 and glucocorticoids as effectors of the resolution of inflammation. Nat Rev Immunol. 2009;9:62–70. https://doi.org/10.1038/nri2470.
D’Acquisto F, Piras G, Rattazzi L. Pro-inflammatory and pathogenic properties of Annexin-A1: the whole is greater than the sum of its parts. Biochem Pharmacol. 2013;85:1213–8. https://doi.org/10.1016/j.bcp.2013.02.011.
Boudhraa Z, Bouchon B, Viallard C, D’Incan M, Degoul F. Annexin A1 localization and its relevance to cancer. Clin Sci. 2016;130:205–20. https://doi.org/10.1042/CS20150415.
Ji AL, Rubin AJ, Thrane K, Jiang S, Reynolds DL, Meyers RM, et al. Multimodal analysis of composition and spatial architecture in human squamous cell carcinoma. Cell. 2020;182:497–514.e422. https://doi.org/10.1016/j.cell.2020.05.039.
Zhou Y, Bian X, Le Y, Gong W, Hu J, Zhang X, et al. Formylpeptide receptor FPR and the rapid growth of malignant human gliomas. J Natl Cancer Inst. 2005;97:823–35. https://doi.org/10.1093/jnci/dji142.
Woroniecka K, Chongsathidkiet P, Rhodin K, Kemeny H, Dechant C, Farber SH, et al. T-cell exhaustion signatures vary with tumor type and are severe in Glioblastoma. Clin Cancer Res. 2018;24:4175–86. https://doi.org/10.1158/1078-0432.CCR-17-1846.
Le Naour J, Liu P, Zhao L, Adjemian S, Sztupinszki Z, Taieb J, et al. A TLR3 ligand reestablishes chemotherapeutic responses in the context of FPR1 deficiency. Cancer Discov. 2021;11:408–23. https://doi.org/10.1158/2159-8290.CD-20-0465.
Okada H, Weller M, Huang R, Finocchiaro G, Gilbert MR, Wick W, et al. Immunotherapy response assessment in neuro-oncology: a report of the RANO working group. Lancet Oncol. 2015;16:e534–42. https://doi.org/10.1016/S1470-2045(15)00088-1.
Picelli S, Faridani OR, Bjorklund AK, Winberg G, Sagasser S, Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nature Protocols. 2014;9:171–81. https://doi.org/10.1038/nprot.2014.006.
Yao L, Wang HM, Song YY, Sui GC. BioQueue: a novel pipeline framework to accelerate bioinformatics analysis. Bioinformatics. 2017;33:3286–8. https://doi.org/10.1093/bioinformatics/btx403.
Shugay M, Bagaev DV, Turchaninova MA, Bolotin DA, Britanova OV, Putintseva EV, et al. VDJtools: Unifying post-analysis of T cell receptor repertoires. Plos Comput Biol. 2015;11:ARTN e1004503. https://doi.org/10.1371/journal.pcbi.1004503.
Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–20. https://doi.org/10.1038/nbt.4096.
Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e1821. https://doi.org/10.1016/j.cell.2019.05.031.
Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc. 2020;15:1484–506. https://doi.org/10.1038/s41596-020-0292-x.
Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet Journal. 2011;17:10–2. https://doi.org/10.14806/ej.17.1.200.
Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–60. https://doi.org/10.1038/nmeth.3317.
Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9. https://doi.org/10.1093/bioinformatics/btu638.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. https://doi.org/10.1186/s13059-014-0550-8.
Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–3. https://doi.org/10.1093/bioinformatics/btp101.
Chanput W, Mes JJ, Wichers HJ. THP-1 cell line: an in vitro cell model for immune modulation approach. Int Immunopharmacol. 2014;23:37–45. https://doi.org/10.1016/j.intimp.2014.08.002.
Ma H, Li YN, Song L, Liu R, Li X, Shang Q, et al. Macrophages inhibit adipogenic differentiation of adipose tissue derived mesenchymal stem/stromal cells by producing pro-inflammatory cytokines. Cell Biosci. 2020;10:88. https://doi.org/10.1186/s13578-020-00450-y.
Acknowledgements
This study was supported by a grant from the National Natural Science Foundation of China (81771309, 31930039 and 31821003 to Xin Lin and 82202983 to Haihui Jiang). This study was also supported by grants from the Capital’s Funds for Health Improvement and Research (2020-2-1075 to Yong Cui) and the National Key Research and Development Program of China (2019YFA0508502 to Xin Lin). The authors would like to thank Dr. Youwen He, Department of Immunology, Duke University Medical Center, for his valuable suggestions during this study.
Author information
Authors and Affiliations
Contributions
Conceptualization, XL, S., and YZ; methodology, YZ, HJ, and NY; formal analysis, YZ, HJ, and NY; investigation, YZ, HJ, NY, SS, DH, LJ., J.L and LX; resources, SL, HJ, ML, KY, XR, and YC; data curation, YZ; writing – original draft, YZ; visualization, YZ, HJ, and NY.; supervision, XL, and SL; funding acquisition, XL SL HJ, and YC.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zheng, Y., Jiang, H., Yang, N. et al. Glioma-derived ANXA1 suppresses the immune response to TLR3 ligands by promoting an anti-inflammatory tumor microenvironment. Cell Mol Immunol 21, 47–59 (2024). https://doi.org/10.1038/s41423-023-01110-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41423-023-01110-0