Abstract
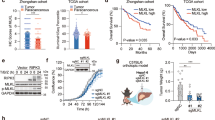
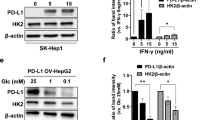
To improve the efficacy of lenvatinib in combination with programmed death-1 (PD-1) blockade therapy for hepatocellular carcinoma (HCC), we screened the suppressive metabolic enzymes that sensitize HCC to lenvatinib and PD-1 blockade, thus impeding HCC progression. After analysis of the CRISPR‒Cas9 screen, phosphatidylinositol-glycan biosynthesis class L (PIGL) ranked first in the positive selection list. PIGL depletion had no effect on tumor cell growth in vitro but reprogrammed the tumor microenvironment (TME) in vivo to support tumor cell survival. Specifically, nuclear PIGL disrupted the interaction between cMyc/BRD4 on the distant promoter of target genes and thus decreased the expression of CCL2 and CCL20, which are involved in shaping the immunosuppressive TME by recruiting macrophages and regulatory T cells. PIGL phosphorylation at Y81 by FGFR2 abolished the interaction of PIGL with importin α/β1, thus retaining PIGL in the cytosol and facilitating tumor evasion by releasing CCL2 and CCL20. Clinically, elevated nuclear PIGL predicts a better prognosis for HCC patients and presents a positive correlation with CD8 + T-cell enrichment in tumors. Clinically, our findings highlight that the nuclear PIGL intensity or the change in PIGL-Y81 phosphorylation should be used as a biomarker to guide lenvatinib with PD-1 blockade therapy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Llovet JM, Castet F, Heikenwalder M, Maini MK, Mazzaferro V, Pinato DJ, et al. Immunotherapies for hepatocellular carcinoma. Nat Rev Clin Oncol. 2022;19:151–72. https://doi.org/10.1038/s41571-021-00573-2.
Makker V, Colombo N, Casado Herraez A, Santin AD, Colomba E, Miller DS, et al. Lenvatinib plus Pembrolizumab for advanced endometrial cancer. N Engl J Med. 2022;386:437–48. https://doi.org/10.1056/NEJMoa2108330.
Labani-Motlagh A, Ashja-Mahdavi M, Loskog A. The tumor microenvironment: A Milieu hindering and obstructing antitumor immune responses. Front Immunol. 2020;11:940 https://doi.org/10.3389/fimmu.2020.00940.
Fukumura D, Kloepper J, Amoozgar Z, Duda DG, Jain RK. Enhancing cancer immunotherapy using antiangiogenics: opportunities and challenges. Nat Rev Clin Oncol. 2018;15:325–40. https://doi.org/10.1038/nrclinonc.2018.29.
Pottekat A, Menon AK. Subcellular localization and targeting of N-acetylglucosaminyl phosphatidylinositol de-N-acetylase, the second enzyme in the glycosylphosphatidylinositol biosynthetic pathway. J Biol Chem. 2004;279:15743–51. https://doi.org/10.1074/jbc.M313537200.
Wang X, Liu R, Zhu W, Chu H, Yu H, Wei P, et al. UDP-glucose accelerates SNAI1 mRNA decay and impairs lung cancer metastasis. Nature. 2019;571:127–31. https://doi.org/10.1038/s41586-019-1340-y.
Yang W, Xia Y, Ji H, Zheng Y, Liang J, Huang W, et al. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature. 2011;480:118–22. https://doi.org/10.1038/nature10598.
Liang C, Shi S, Qin Y, Meng Q, Hua J, Hu Q, et al. Localisation of PGK1 determines metabolic phenotype to balance metastasis and proliferation in patients with SMAD4-negative pancreatic cancer. Gut. 2020;69:888–900. https://doi.org/10.1136/gutjnl-2018-317163.
Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. https://doi.org/10.1016/j.cell.2012.03.003.
Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1–11. https://doi.org/10.1128/MCB.19.1.1.
Morrison-Smith CD, Knox TM, Filic I, Soroko KM, Eschle BK, Wilkens MK, et al. Combined targeting of the BRD4-NUT-p300 axis in NUT Midline carcinoma by dual selective Bromodomain inhibitor, NEO2734. Mol Cancer Ther. 2020;19:1406–14. https://doi.org/10.1158/1535-7163.MCT-20-0087.
Patel MC, Debrosse M, Smith M, Dey A, Huynh W, Sarai N, et al. BRD4 coordinates recruitment of pause release factor P-TEFb and the pausing complex NELF/DSIF to regulate transcription elongation of interferon-stimulated genes. Mol Cell Biol. 2013;33:2497–507. https://doi.org/10.1128/MCB.01180-12.
Muhar M, Ebert A, Neumann T, Umkehrer C, Jude J, Wieshofer C, et al. SLAM-seq defines direct gene-regulatory functions of the BRD4-MYC axis. Science. 2018;360:800–5. https://doi.org/10.1126/science.aao2793.
Dang CV, Reddy EP, Shokat KM, Soucek L. Drugging the ‘undruggable’ cancer targets. Nat Rev Cancer. 2017;17:502–8. https://doi.org/10.1038/nrc.2017.36.
Chen H, Liu H, Qing G. Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduct Target Ther. 2018;3:5 https://doi.org/10.1038/s41392-018-0008-7.
Llombart V, Mansour MR. Therapeutic targeting of “undruggable” MYC. EBioMedicine. 2022;75:103756 https://doi.org/10.1016/j.ebiom.2021.103756.
Scharping NE, Menk AV, Moreci RS, Whetstone RD, Dadey RE, Watkins SC, et al. The tumor microenvironment represses T cell mitochondrial biogenesis to drive intratumoral T Cell metabolic insufficiency and dysfunction. Immunity. 2016;45:374–88. https://doi.org/10.1016/j.immuni.2016.07.009.
Gutzmer R, Stroyakovskiy D, Gogas H, Robert C, Lewis K, Protsenko S, et al. Atezolizumab, vemurafenib, and cobimetinib as first-line treatment for unresectable advanced BRAF(V600) mutation-positive melanoma (IMspire150): Primary analysis of the randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2020;395:1835–44. https://doi.org/10.1016/S0140-6736(20)30934-X.
Rodell CB, Arlauckas SP, Cuccarese MF, Garris CS, Li R, Ahmed MS, et al. TLR7/8-agonist-loaded nanoparticles promote the polarization of tumour-associated macrophages to enhance cancer immunotherapy. Nat Biomed Eng. 2018;2:578–88. https://doi.org/10.1038/s41551-018-0236-8.
Zhu Y, Yang J, Xu D, Gao XM, Zhang Z, Hsu JL, et al. Disruption of tumour-associated macrophage trafficking by the osteopontin-induced colony-stimulating factor-1 signalling sensitises hepatocellular carcinoma to anti-PD-L1 blockade. Gut. 2019;68:1653–66. https://doi.org/10.1136/gutjnl-2019-318419.
Xu D, Shao F, Bian X, Meng Y, Liang T, Lu Z. The evolving landscape of noncanonical functions of metabolic enzymes in cancer and other pathologies. Cell Metab. 2021;33:33–50. https://doi.org/10.1016/j.cmet.2020.12.015.
Luo W, Hu H, Chang R, Zhong J, Knabel M, O'Meally R, et al. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145:732–44. https://doi.org/10.1016/j.cell.2011.03.054.
Rahl PB, Young RA. MYC and transcription elongation. Cold Spring Harb Perspect Med. 2014;4:a020990 https://doi.org/10.1101/cshperspect.a020990.
Durbin AD, Zimmerman MW, Dharia NV, Abraham BJ, Iniguez AB, Weichert-Leahey N, et al. Selective gene dependencies in MYCN-amplified neuroblastoma include the core transcriptional regulatory circuitry. Nat Genet. 2018;50:1240–6. https://doi.org/10.1038/s41588-018-0191-z.
Schoenfelder S, Fraser P. Long-range enhancer-promoter contacts in gene expression control. Nat Rev Genet. 2019;20:437–55. https://doi.org/10.1038/s41576-019-0128-0.
Morton AR, Dogan-Artun N, Faber ZJ, MacLeod G, Bartels CF, Piazza MS, et al. Functional enhancers shape extrachromosomal oncogene amplifications. Cell. 2019;179:1330–1341 e1313. https://doi.org/10.1016/j.cell.2019.10.039.
Alekseyenko AA, Walsh EM, Wang X, Grayson AR, Hsi PT, Kharchenko PV, et al. The oncogenic BRD4-NUT chromatin regulator drives aberrant transcription within large topological domains. Genes Dev. 2015;29:1507–23. https://doi.org/10.1101/gad.267583.115.
Roussos ET, Condeelis JS, Patsialou A. Chemotaxis in cancer. Nat Rev Cancer. 2011;11:573–87. https://doi.org/10.1038/nrc3078.
Sahin H, Trautwein C, Wasmuth HE. Functional role of chemokines in liver disease models. Nat Rev Gastroenterol Hepatol. 2010;7:682–90. https://doi.org/10.1038/nrgastro.2010.168.
Mandrekar P, Ambade A, Lim A, Szabo G, Catalano D. An essential role for monocyte chemoattractant protein-1 in alcoholic liver injury: regulation of proinflammatory cytokines and hepatic steatosis in mice. Hepatology. 2011;54:2185–97. https://doi.org/10.1002/hep.24599.
Zhang J, Patel L, Pienta KJ. Targeting chemokine (C-C motif) ligand 2 (CCL2) as an example of translation of cancer molecular biology to the clinic. Prog Mol Biol Transl Sci. 2010;95:31–53. https://doi.org/10.1016/B978-0-12-385071-3.00003-4.
Qian BZ, Li J, Zhang H, Kitamura T, Zhang J, Campion LR, et al. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475:222–5. https://doi.org/10.1038/nature10138.
Pienta KJ, Machiels JP, Schrijvers D, Alekseev B, Shkolnik M, Crabb SJ, et al. Phase 2 study of carlumab (CNTO 888), a human monoclonal antibody against CC-chemokine ligand 2 (CCL2), in metastatic castration-resistant prostate cancer. Invest N Drugs. 2013;31:760–8. https://doi.org/10.1007/s10637-012-9869-8.
Benkheil M, Van Haele M, Roskams T, Laporte M, Noppen S, Abbasi K, et al. CCL20, a direct-acting pro-angiogenic chemokine induced by hepatitis C virus (HCV): Potential role in HCV-related liver cancer. Exp Cell Res. 2018;372:168–77. https://doi.org/10.1016/j.yexcr.2018.09.023.
Cook KW, Letley DP, Ingram RJ, Staples E, Skjoldmose H, Atherton JC, et al. CCL20/CCR6-mediated migration of regulatory T cells to the Helicobacter pylori-infected human gastric mucosa. Gut. 2014;63:1550–9. https://doi.org/10.1136/gutjnl-2013-306253.
Lee B, Namkoong H, Yang Y, Huang H, Heller D, Szot GL, et al. Single-cell sequencing unveils distinct immune microenvironments with CCR6-CCL20 crosstalk in human chronic pancreatitis. Gut. 2022;71:1831–42. https://doi.org/10.1136/gutjnl-2021-324546.
Kawanabe-Matsuda H, Takeda K, Nakamura M, Makino S, Karasaki T, Kakimi K, et al. Dietary Lactobacillus-derived exopolysaccharide enhances immune-checkpoint blockade therapy. Cancer Discov. 2022;12:1336–55. https://doi.org/10.1158/2159-8290.CD-21-0929.
Joung J, Konermann S, Gootenberg JS, Abudayyeh OO, Platt RJ, Brigham MD, et al. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat Protoc. 2017;12:828–63. https://doi.org/10.1038/nprot.2017.016.
Wang B, Wang M, Zhang W, Xiao T, Chen CH, Wu A, et al. Integrative analysis of pooled CRISPR genetic screens using MAGeCKFlute. Nat Protoc. 2019;14:756–80. https://doi.org/10.1038/s41596-018-0113-7.
Chen P, Luo X, Dai G, Jiang Y, Luo Y, Peng S, et al. Dexmedetomidine promotes the progression of hepatocellular carcinoma through hepatic stellate cell activation. Exp Mol Med. 2020;52:1062–74. https://doi.org/10.1038/s12276-020-0461-6.
Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16:1289–96. https://doi.org/10.1038/s41592-019-0619-0.
Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. 2019;20:163–72. https://doi.org/10.1038/s41590-018-0276-y.
Yu G, Wang LG, Han Y, He QY. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–7. https://doi.org/10.1089/omi.2011.0118.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50. https://doi.org/10.1073/pnas.0506580102.
Acknowledgements
The authors thank the Centre for Precise Gene Editing, Guangzhou University for technical support.
Funding
This study was supported mainly by the Guangdong Natural Science Foundation of Guangdong Province of China 2021B1515020016 and in part by NSFC grants No. 82000616, 82272714, 82173149 and Science and Technology Program of Guangdong Province No. 2020B1212060019.
Author information
Authors and Affiliations
Contributions
HY, TZS, LLY, and DWX designed, performed, and analyzed the experiments and prepared the figures. WL and QX provided technical and clinical assistance. XJW and YFD conceived this study and wrote the manuscript, and XJW, WL, and QX revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declared no competing interests.
Ethical approval
The research content and implementation plan of this experiment strictly complied with the Declaration of Helsinki and was approved by the ethics committee of The Third Affiliated Hospital of Sun Yat-sen University (Medical Ethics 2022-02-333-01).
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yu, H., Shi, T., Yao, L. et al. Elevated nuclear PIGL disrupts the cMyc/BRD4 axis and improves PD-1 blockade therapy by dampening tumor immune evasion. Cell Mol Immunol 20, 867–880 (2023). https://doi.org/10.1038/s41423-023-01048-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41423-023-01048-3
Keywords
This article is cited by
-
CRISPR–Cas9 applications in T cells and adoptive T cell therapies
Cellular & Molecular Biology Letters (2024)
-
Mechanism-directed combinational immunotherapies in liver cancer hold promise
Cellular & Molecular Immunology (2023)