Abstract
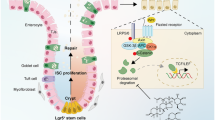
Dysregulation of gut homeostasis is associated with irritable bowel syndrome (IBS), a chronic functional gastrointestinal disorder affecting approximately 11.2% of the global population. The poorly understood pathogenesis of IBS has impeded its treatment. Here, we report that the E3 ubiquitin ligase tripartite motif-containing 27 (TRIM27) is weakly expressed in IBS but highly expressed in inflammatory bowel disease (IBD), a frequent chronic organic gastrointestinal disorder. Accordingly, knockout of Trim27 in mice causes spontaneously occurring IBS-like symptoms, including increased visceral hyperalgesia and abnormal stool features, as observed in IBS patients. Mechanistically, TRIM27 stabilizes β-catenin and thus activates Wnt/β-catenin signaling to promote intestinal stem cell (ISC) self-renewal. Consistent with these findings, Trim27 deficiency disrupts organoid formation, which is rescued by reintroducing TRIM27 or β-catenin. Furthermore, Wnt/β-catenin signaling activator treatment ameliorates IBS symptoms by promoting ISC self-renewal. Taken together, these data indicate that TRIM27 is critical for maintaining gut homeostasis, suggesting that targeting the TRIM27/Wnt/β-catenin axis could be a potential treatment strategy for IBS. Our study also indicates that TRIM27 might serve as a potential biomarker for differentiating IBS from IBD.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout









Similar content being viewed by others
Data availability
The 16S rRNA gene sequences are available in the NCBI Sequence Read Archive (SRA) database under accession number SRP355336. Publicly available microarray data (E-MTAB-5811, GSE1710) were downloaded from the EMBL-EBI ArrayExpress database and NCBI Gene Expression Omnibus. Any additional information required to reanalyze the data reported in this paper is available from the corresponding author upon request.
References
Sperber AD. Epidemiology and burden of irritable bowel syndrome: an international perspective. Gastroenterol Clin North Am. 2021;50:489–503.
Ng SC, Shi HY, Hamidi N, Underwood FE, Tang W, Benchimol EI, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. 2017;390:2769–78.
Kaplan GG, Windsor JW. The four epidemiological stages in the global evolution of inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2021;18:56–66.
Hanning N, Edwinson AL, Ceuleers H, Peters SA, De Man JG, Hassett LC, et al. Intestinal barrier dysfunction in irritable bowel syndrome: a systematic review. Ther Adv Gastroenterol. 2021;14:1756284821993586.
Spiller R, Major G. IBS and IBD – separate entities or on a spectrum? Nat Rev Gastroenterol Hepatol. 2016;13:613–21.
Santos AJM, Lo YH, Mah AT, Kuo CJ. The intestinal stem cell niche: homeostasis and adaptations. Trends Cell Biol. 2018;28:1062–78.
Ahlawat S, Kumar P, Mohan H, Goyal S, Sharma KK. Inflammatory bowel disease: tri-directional relationship between microbiota, immune system and intestinal epithelium. Crit Rev Microbiol. 2021;47:254–73.
Eijsbouts C, Zheng T, Kennedy NA, Bonfiglio F, Anderson CA, Moutsianas L, et al. Genome-wide analysis of 53,400 people with irritable bowel syndrome highlights shared genetic pathways with mood and anxiety disorders. Nat Genet. 2021;53:1543–52.
Zhu L, Li Y, Zhou L, Yang G, Wang Y, Han J, et al. Role of RING-Type E3 ubiquitin ligases in inflammatory signalling and inflammatory bowel disease. Mediators Inflamm. 2020;2020:5310180.
Cai X, Luo Y, Zhang Y, Lin Y, Wu B, Cao Z, et al. The deubiquitinase OTUD1 inhibits colonic inflammation by suppressing RIPK1-mediated NF-kappaB signaling. Cell Mol Immunol. 2022;19:276–89.
Chen SY, Zhang HP, Li J, Shi JH, Tang HW, Zhang Y, et al. Tripartite motif-containing 27 attenuates liver ischemia/reperfusion injury by suppressing transforming growth factor beta-activated kinase 1 (TAK1) by TAK1 binding protein 2/3 degradation. Hepatology. 2021;73:738–58.
Gushchina LV, Kwiatkowski TA, Bhattacharya S, Weisleder NL. Conserved structural and functional aspects of the tripartite motif gene family point towards therapeutic applications in multiple diseases. Pharm Ther. 2018;185:12–25.
Zurek B, Schoultz I, Neerincx A, Napolitano LM, Birkner K, Bennek E, et al. TRIM27 negatively regulates NOD2 by ubiquitination and proteasomal degradation. PLoS One. 2012;7:e41255.
Zheng Q, Hou J, Zhou Y, Yang Y, Xie B, Cao X. Siglec1 suppresses antiviral innate immune response by inducing TBK1 degradation via the ubiquitin ligase TRIM27. Cell Res. 2015;25:1121–36.
Zhang HX, Xu ZS, Lin H, Li M, Xia T, Cui K, et al. TRIM27 mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis. Nat Commun. 2018;9:3441.
Wang R, Li H, Wu J, Cai ZY, Li B, Ni H, et al. Gut stem cell necroptosis by genome instability triggers bowel inflammation. Nature. 2020;580:386–90.
Hong KB, Seo H, Lee JS, Park Y. Effects of probiotic supplementation on post-infectious irritable bowel syndrome in rodent model. BMC Complement Alter Med. 2019;19:195.
Kodani M, Fukui H, Tomita T, Oshima T, Watari J, Miwa H. Association between gastrointestinal motility and macrophage/mast cell distribution in mice during the healing stage after DSSinduced colitis. Mol Med Rep. 2018;17:8167–72.
Wilkinson JM, Gill MC. Irritable bowel syndrome: questions and answers for effective care. Am Fam Physician. 2021;103:727–36.
Zhou XY. Visceral hypersensitive rats share common dysbiosis features with irritable bowel syndrome patients. World J Gastroenterol. 2016;22:5211–27.
Zielinska M, Fichna J, Bashashati M, Habibi S, Sibaev A, Timmermans JP, et al. G protein-coupled estrogen receptor and estrogen receptor ligands regulate colonic motility and visceral pain. Neurogastroenterol Motil. 2017;29:e13025.
Grabauskas G, Wu X, Gao J, Li JY, Turgeon DK, Owyang C. Prostaglandin E2, produced by mast cells in colon tissues from patients with irritable bowel syndrome, contributes to visceral hypersensitivity in mice. Gastroenterology. 2020;158:2195–207.e2196.
Pimentel M, Lembo A. Microbiome and its role in irritable bowel syndrome. Dig Dis Sci. 2020;65:829–39.
Blonska A, Konrad P, Chojnacki J, Chojnacki C. [Evaluation of oro-cecal transit time in patients with irritable bowel syndrome with cereal products intolerance]. Pol Merkur Lekarski. 2017;42:116–20.
Yoshimoto T, Oshima T, Huang X, Tomita T, Fukui H, Miwa H. Microinflammation in the intestinal mucosa and symptoms of irritable bowel syndrome. J Gastroenterol. 2021;57:62–69.
Uranga JA, Martinez V, Abalo R. Mast cell regulation and irritable bowel syndrome: effects of food components with potential nutraceutical use. Molecules. 2020;25:4314.
Bazzoni G, Martınez-Estrada OM, Orsenigo F, Cordenonsi M, Citi S, Dejana E. Interaction of junctional adhesion molecule with the tight junction components ZO-1, cingulin, and occludin. J Biol Chem. 2000;275:20520–6.
Linsalata M, Riezzo G, Clemente C, D’Attoma B, Russo F. Noninvasive biomarkers of gut barrier function in patients suffering from diarrhea predominant-IBS: an update. Dis Markers. 2020;2020:2886268.
Vich Vila A, Imhann F, Collij V, Jankipersadsing SA, Gurry T, Mujagic Z, et al. Gut microbiota composition and functional changes in inflammatory bowel disease and irritable bowel syndrome. Sci Transl Med. 2018;10:eaap8914.
Vervier K, Moss S, Kumar N, Adoum A, Barne M, Browne H, et al. Two microbiota subtypes identified in irritable bowel syndrome with distinct responses to the low FODMAP diet. Gut. 2021;71:1821–30.
Fritz T, Niederreiter L, Adolph T, Blumberg RS, Kaser A. Crohn’s disease: NOD2, autophagy and ER stress converge. Gut. 2011;60:1580–8.
Lee SM, Kim N, Yoon H, Kim YS, Choi SI, Park JH, et al. Compositional and functional changes in the gut microbiota in irritable bowel syndrome patients. Gut Liver. 2021;15:253–61.
Metcalfe C, Kljavin NM, Ybarra R, de Sauvage FJ. Lgr5+ stem cells are indispensable for radiation-induced intestinal regeneration. Cell Stem Cell. 2014;14:149–59.
Farin HF, Jordens I, Mosa MH, Basak O, Korving J, Tauriello DVF, et al. Visualization of a short-range Wnt gradient in the intestinal stem-cell niche. Nature. 2016;530:340–3.
Zha JM, Li HS, Lin Q, Kuo WT, Jiang ZH, Tsai PY, et al. Interleukin 22 expands transit-amplifying cells while depleting Lgr5(+) stem cells via inhibition of Wnt and Notch signaling. Cell Mol Gastroenterol Hepatol. 2019;7:255–74.
Nusse R, Clevers H. Wnt/beta-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169:985–99.
Ikeda S. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3beta and beta-catenin and promotes GSK-3beta-dependent phosphorylation of beta-catenin. EMBO J. 1998;17:1371–84.
Hasegawa N, Iwashita T, Asai N, Murakami H, Iwata Y, Isomura T, et al. A RING finger motif regulates transforming activity of the rfp/ret fusion gene. Biochem Biophys Res Commun. 1996;225:627–31.
Gwak J, Hwang SG, Park HS, Choi SR, Park SH, Kim H, et al. Small molecule-based disruption of the Axin/beta-catenin protein complex regulates mesenchymal stem cell differentiation. Cell Res. 2012;22:237–47.
Delgado S, Sanchez B, Margolles A, Ruas-Madiedo P, Ruiz L. Molecules produced by probiotics and intestinal microorganisms with immunomodulatory activity. Nutrients. 2020;12:391.
Akutko K, Stawarski A. Probiotics, prebiotics and synbiotics in inflammatory bowel diseases. J Clin Med. 2021;10:2466.
Szalwinska P, Wlodarczyk J, Spinelli A, Fichna J, Wlodarczyk M. IBS-symptoms in IBD patients-manifestation of concomitant or different entities. J Clin Med. 2020;10:31.
Johnson AC, Farmer AD, Ness TJ, Greenwood-Van Meerveld B. Critical evaluation of animal models of visceral pain for therapeutics development: a focus on irritable bowel syndrome. Neurogastroenterol Motil. 2020;32:e13776.
Dotti I, Mora-Buch R, Ferrer-Picon E, Planell N, Jung P, Masamunt MC, et al. Alterations in the epithelial stem cell compartment could contribute to permanent changes in the mucosa of patients with ulcerative colitis. Gut. 2017;66:2069–79.
Du G, Xiong L, Li X, Zhuo Z, Zhuang X, Yu Z, et al. Peroxisome elevation induces stem cell differentiation and intestinal epithelial repair. Dev Cell. 2020;53:169–84. e111
Fredericks E, Dealtry G, Roux S. beta-Catenin regulation in sporadic colorectal carcinogenesis: not as simple as APC. Can J Gastroenterol Hepatol. 2018;2018:4379673.
Keller DS, Windsor A, Cohen R, Chand M. Colorectal cancer in inflammatory bowel disease: review of the evidence. Tech Coloproctol. 2019;23:3–13.
Wu S, Yuan C, Liu S, Zhang Q, Yang Z, Sun F, et al. Irritable bowel syndrome and long-term risk of cancer: a prospective cohort study among 0.5 million adults in UK Biobank. Am J Gastroenterol. 2022;117:785–93.
Schmitt M, Schewe M, Sacchetti A, Feijtel D, van de Geer WS, Teeuwssen M, et al. Paneth cells respond to inflammation and contribute to tissue regeneration by acquiring stem-like features through SCF/c-Kit signaling. Cell Rep. 2018;24:2312–28.e2317.
Zhang H, Lin M, Dong C, Tang Y, An L, Ju J, et al. An MST4-pbeta-Catenin(Thr40) signaling axis controls intestinal stem cell and tumorigenesis. Adv Sci (Weinh). 2021;8:e2004850.
Kayama H, Okumura R, Takeda K. Interaction between the microbiota, epithelia, and immune cells in the intestine. Annu Rev Immunol. 2020;38:23–48.
Neurath MF. Targeting immune cell circuits and trafficking in inflammatory bowel disease. Nat Immunol. 2019;20:970–9.
Mars RAT, Yang Y, Ward T, Houtti M, Priya S, Lekatz HR, et al. Longitudinal multi-omics reveals subset-specific mechanisms underlying irritable bowel syndrome. Cell. 2020;182:1460–73.e1417.
Ford AC, Sperber AD, Corsetti M, Camilleri M. Irritable bowel syndrome. Lancet. 2020;396:1675–88.
Kumar S, Singh P, Kumar A. Targeted therapy of irritable bowel syndrome with anti-inflammatory cytokines. Clin J Gastroenterol. 2021;15:1–10.
Enck P, Aziz Q, Barbara G, Farmer AD, Fukudo S, Mayer EA, et al. Irritable bowel syndrome. Nat Rev Dis Prim. 2016;2:16014.
Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–7.
Lu Y, Huang J, Zhang Y, Huang Z, Yan W, Zhou T, et al. Therapeutic effects of berberine hydrochloride on stress-induced diarrhea-predominant irritable bowel syndrome rats by inhibiting neurotransmission in colonic smooth muscle. Front Pharm. 2021;12:596686.
Wang J, Li BX, Ge PP, Li J, Wang Q, Gao GF, et al. Mycobacterium tuberculosis suppresses innate immunity by coopting the host ubiquitin system. Nat Immunol. 2015;16:237–45.
Gautier L, Cope L, Bolstad BM, Irizarry RA. affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–15.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47.
Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–25.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50.
Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–73.
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6.
Rognes T, Flouri T, Nichols B, Quince C, Mahe F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584.
Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60.
Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, et al. QuPath: Open source software for digital pathology image analysis. Sci Rep. 2017;7:16878.
Acknowledgements
We thank Junfeng Hao (Core Facility for Protein Research, Institute of Biophysics, Chinese Academy of Sciences, Beijing) for helping with the histological analysis and Zhihua Liu and Hongying Wang (National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College) for helping with the endoscopy experiment. This work was supported by the National Key Research and Development Project of China (2021YFA1300200 to CHL and LZ, 2022YFC2302900 to CHL and JW), the National Natural Science Foundation of China (81825014 to CHL, 31830003 to CHL, 82022041 to JW and 81871616 to JW), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB29020000 to CHL), Youth Innovation Promotion Association CAS (2018118 to JW) and the State Key Laboratory of Proteomics (SKLP-K202001 to LZ and SKLPO202003 to JW).
Author information
Authors and Affiliations
Contributions
CHL and LZ conceived and supervised the study. CHL and JW designed the experiments. JW, DZ and PG performed the majority of the experiments. ZLei, ZLu, QC, YZ, LQ, YY, XZ, BL and SZ helped with some of the experiments and data analysis. CHL, JW and ZLei analyzed the data and wrote the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, J., Zhao, D., Lei, Z. et al. TRIM27 maintains gut homeostasis by promoting intestinal stem cell self-renewal. Cell Mol Immunol 20, 158–174 (2023). https://doi.org/10.1038/s41423-022-00963-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41423-022-00963-1
Keywords
This article is cited by
-
Intestinal stem cells: guardians of homeostasis in health and aging amid environmental challenges
Experimental & Molecular Medicine (2024)