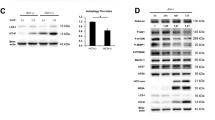
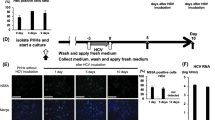
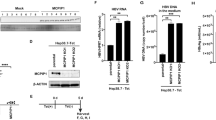
Abstract
HBV is considered as a “stealth” virus that does not invoke interferon (IFN) responses; however, the mechanisms by which HBV bypasses innate immune recognition are poorly understood. In this study, we identified adenosine deaminases acting on RNA 1 (ADAR1), which is a key factor in HBV evasion from IFN responses in hepatocytes. Mechanically, ADAR1 interacted with HBV RNAs and deaminated adenosine (A) to generate inosine (I), which disrupted host immune recognition and thus promoted HBV replication. Loss of ADAR1 or its deficient deaminase activity promoted IFN responses and inhibited HBV replication in hepatocytes, and blocking the IFN signaling pathways released the inhibition of HBV replication caused by ADAR1 deficiency. Notably, the HBV X protein (HBx) transcriptionally promoted ADAR1 expression to increase the threshold required to trigger intrinsic immune activation, which in turn enhanced HBV escape from immune recognition, leading to persistent infection. Supplementation with 8-azaadenosine, an ADAR1 inhibitor, efficiently enhanced liver immune activation to promote HBV clearance in vivo and in vitro. Taken together, our results delineate a molecular mechanism by which HBx promotes ADAR1-derived HBV immune escape and suggest a targeted therapeutic intervention for HBV infection.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Seeger C. Control of viral transcripts as a concept for future HBV therapies. Curr Opin Virol. 2018;30:18–23.
Dansako H, Ueda Y, Okumura N, Satoh S, Sugiyama M, Mizokami M, et al. The cyclic GMP-AMP synthetase-STING signaling pathway is required for both the innate immune response against HBV and the suppression of HBV assembly. FEBS J. 2016;283:144–56.
Stacey AR, Norris PJ, Qin L, Haygreen EA, Taylor E, Heitman J, et al. Induction of a striking systemic cytokine cascade prior to peak viremia in acute human immunodeficiency virus type 1 infection, in contrast to more modest and delayed responses in acute hepatitis B and C virus infections. J Virol. 2009;83:3719–33.
Wieland S, Thimme R, Purcell RH, Chisari FV. Genomic analysis of the host response to hepatitis B virus infection. Proc Natl Acad Sci USA. 2004;101:6669–74.
Mutz P, Metz P, Lempp FA, Bender S, Qu B, Schöneweis K, et al. HBV bypasses the innate immune response and does not protect HCV from antiviral activity of interferon. Gastroenterology. 2018;154:1791–804.
Wei C, Ni C, Song T, Liu Y, Yang X, Zheng Z, et al. The hepatitis B virus X protein disrupts innate immunity by downregulating mitochondrial antiviral signaling protein. J Immunol. 2010;185:1158–68.
Luangsay S, Gruffaz M, Isorce N, Testoni B, Michelet M, Faure-Dupuy S, et al. Early inhibition of hepatocyte innate responses by hepatitis B virus. J Hepatol. 2015;63:1314–22.
Sato S, Li K, Kameyama T, Hayashi T, Ishida Y, Murakami S, et al. The RNA sensor RIG-I dually functions as an innate sensor and direct antiviral factor for hepatitis B virus. Immunity. 2015;42:123–32.
Thomsen MK, Nandakumar R, Stadler D, Malo A, Valls RM, Wang F, et al. Lack of immunological DNA sensing in hepatocytes facilitates hepatitis B virus infection. Hepatology. 2016;64:746–59.
Cheng X, Xia Y, Serti E, Block PD, Chung M, Chayama K, et al. Hepatitis B virus evades innate immunity of hepatocytes but activates cytokine production by macrophages. Hepatology. 2017;66:1779–93.
Suslov A, Boldanova T, Wang X, Wieland S, Heim MH. Hepatitis B virus does not interfere with innate immune responses in the human liver. Gastroenterology. 2018;154:1778–90.
Li K, Chen Z, Kato N, Gale M Jr., Lemon SM. Distinct poly(I-C) and virus-activated signaling pathways leading to interferon-beta production in hepatocytes. J Biol Chem Iological Chem. 2005;280:16739–47.
Yin X, Li X, Ambardekar C, Hu Z, Lhomme S, Feng Z. Hepatitis E virus persists in the presence of a type III interferon response. PLoS Pathog. 2017;13:e1006417.
Sayed IM, Verhoye L, Cocquerel L, Abravanel F, Foquet L, Montpellier C, et al. Study of hepatitis E virus infection of genotype 1 and 3 in mice with humanised liver. Gut. 2017;66:920–9.
Freund I, Eigenbrod T, Helm M, Dalpke AH. RNA modifications modulate activation of innate Toll-like receptors. Genes. 2019;10:92.
Mannion NM, Greenwood SM, Young R, Cox S, Brindle J, Read D, et al. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep. 2014;9:1482–94.
Pestal K, Funk CC, Snyder JM, Price ND, Treuting PM, Stetson DB. Isoforms of RNA-editing enzyme ADAR1 independently control nucleic acid sensor MDA5-driven autoimmunity and multi-organ development. Immunity. 2015;43:933–44.
Liddicoat BJ, Piskol R, Chalk AM, Ramaswami G, Higuchi M, Hartner JC, et al. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science. 2015;349:1115–20.
Yang S, Deng P, Zhu Z, Zhu J, Wang G, Zhang L, et al. Adenosine deaminase acting on RNA 1 limits RIG-I RNA detection and suppresses IFN production responding to viral and endogenous RNAs. J Immunol. 2014;193:3436–45.
Pfaller CK, Donohue RC, Nersisyan S, Brodsky L, Cattaneo R. Extensive editing of cellular and viral double-stranded RNA structures accounts for innate immunity suppression and the proviral activity of ADAR1p150. PLoS Biol. 2018;16:e2006577.
Pujantell M, Franco S, Galván-Femenía I, Badia R, Castellví M, Garcia-Vidal E, et al. ADAR1 affects HCV infection by modulating innate immune response. Antivir Res. 2018;156:116–27.
Yang D, Zuo C, Wang X, Meng X, Xue B, Liu N, et al. Complete replication of hepatitis B virus and hepatitis C virus in a newly developed hepatoma cell line. Proc Natl Acad Sci USA. 2014;111:E1264–73.
Duriez M, Mandouri Y, Lekbaby B, Wang H, Schnuriger A, Redelsperger F, et al. Alternative splicing of hepatitis B virus: a novel virus/host interaction altering liver immunity. J Hepatol. 2017;67:687–99.
Thomas JM, Beal PA. How do ADARs bind RNA? New protein-RNA structures illuminate substrate recognition by the RNA editing ADARs. BioEssays. 2017;39:10.
Jeong JK, Yoon GS, Ryu WS. Evidence that the 5’-end cap structure is essential for encapsidation of hepatitis B virus pregenomic RNA. J Virol. 2000;74:5502–8.
Gallo A, Locatelli F. ADARs: allies or enemies? The importance of A-to-I RNA editing in human disease: from cancer to HIV-1. Biol Rev Camb Philos Soc. 2012;87:95–110.
Valente L, Nishikura K. RNA binding-independent dimerization of adenosine deaminases acting on RNA and dominant negative effects of nonfunctional subunits on dimer functions. J Biol Chem. 2007;282:16054–61.
Pujantell M, Riveira-Muñoz E, Badia R, Castellví M, Garcia-Vidal E, Sirera G, et al. RNA editing by ADAR1 regulates innate and antiviral immune functions in primary macrophages. Sci Rep. 2017;7:13339.
Li T, Yang X, Li W, Song J, Li Z, Zhu X, et al. ADAR1 stimulation by IFN-alpha downregulates the expression of MAVS via RNA editing to regulate the anti-HBV response. Mol Ther. 2021;29:1335–48.
Leong CR, Oshiumi H, Suzuki T, Matsumoto M, Seya T. Nucleic acid sensors involved in the recognition of HBV in the liver-specific in vivo transfection mouse models-pattern recognition receptors and sensors for HBV. Med Sci. 2015;3:16–24.
Tomaselli S, Galeano F, Locatelli F, Gallo A. ADARs and the balance game between virus infection and innate immune cell response. Curr Issues Mol Biol. 2015;17:37–51.
Doria M, Neri F, Gallo A, Farace MG, Michienzi A. Editing of HIV-1 RNA by the double-stranded RNA deaminase ADAR1 stimulates viral infection. Nucleic Acids Res. 2009;37:5848–58.
Yang Y, Zhou X, Jin Y. ADAR-mediated RNA editing in non-coding RNA sequences. Sci China Life Sci. 2013;56:944–52.
Barraud P, Banerjee S, Mohamed W, Jantsch M, Allain F. A bimodular nuclear localization signal assembled via an extended double-stranded RNA-binding domain acts as an RNA-sensing signal for transportin 1. Proc Natl Acad Sci USA. 2014;111:E1852–61.
Guo X, Chen P, Hou X, Xu W, Wang D, Wang TY, et al. The recombined cccDNA produced using minicircle technology mimicked HBV genome in structure and function closely. Sci Rep. 2016;6:25552.
Zipeto MA, Court AC, Sadarangani A, Delos Santos NP, Balaian L, Chun HJ, et al. ADAR1 activation drives leukemia stem cell self-renewal by impairing let-7 biogenesis. Cell Stem Cell. 2016;19:177–91.
Xu L, Wu Z, Tan S, Wang Z, Lin Q, Li X, et al. Tumor suppressor ZHX2 restricts hepatitis B virus replication via epigenetic and non-epigenetic manners. Antivir Res. 2018;153:114–23.
Liu Y, Li J, Chen J, Li Y, Wang W, Du X, et al. Hepatitis B virus polymerase disrupts K63-linked ubiquitination of STING to block innate cytosolic DNA-sensing pathways. J Virol. 2015;89:2287–300.
Jiang J, Tang H. Mechanism of inhibiting type I interferon induction by hepatitis B virus X protein. Protein Cell. 2010;1:1106–17.
Wang G, Wang H, Singh S, Zhou P, Yang S, Wang Y, et al. ADAR1 prevents liver injury from inflammation and suppresses interferon production in hepatocytes. Am J Pathol. 2015;185:3224–37.
Wang H, Wang G, Zhang L, Zhang J, Zhang J, Wang Q, et al. ADAR1 suppresses the activation of cytosolic RNA-sensing signaling pathways to protect the liver from ischemia/reperfusion injury. Sci Rep. 2016;6:20248.
Lu HL, Liao F. Melanoma differentiation-associated gene 5 senses hepatitis B virus and activates innate immune signaling to suppress virus replication. J Immunol. 2013;191:3264–76.
Chung H, Calis JJA, Wu X, Sun T, Yu Y, Sarbanes SL, et al. Human ADAR1 prevents endogenous RNA from triggering translational shutdown. Cell. 2018;172:811–24.
Liu G, Ma X, Wang Z, Wakae K, Yuan Y, He Z, et al. Adenosine deaminase acting on RNA-1 (ADAR1) inhibits hepatitis B virus (HBV) replication by enhancing microRNA-122 processing. J Biol Chem. 2019;294:14043–54.
Yuan L, Jia Q, Yang S, Idris N, Li Y, Wang Y, et al. ADAR1 promotes HBV replication through its deaminase domain. Front Biosci. 2020;25:710–21.
Sung WK, Lu Y, Lee C, Zhang D, Ronaghi M, Lee C. Deregulated direct targets of the hepatitis B virus (HBV) protein, HBx, identified through chromatin immunoprecipitation and expression microarray profiling. J Biol Chem. 2009;284:21941–54.
Shen C, Feng X, Mao T, Yang D, Zou J, Zao X, et al. Yin-Yang 1 and HBx protein activate HBV transcription by mediating the spatial interaction of cccDNA minichromosome with cellular chromosome 19p13.11. Emerg Microbes Infect. 2020;9:2455–64.
Shan X, Ren M, Chen K, Huang A, Tang H. Regulation of the microRNA processor DGCR8 by hepatitis B virus proteins via the transcription factor YY1-803. Arch Virol. 2015;160:795–803.
Zhang L, Cai X, Chen K, Wang Z, Wang L, Ren M, et al. Hepatitis B virus protein up-regulated HLJ1 expression via the transcription factor YY1 in human hepatocarcinoma cells. Virus Res. 2011;157:76–81.
Ishizuka JJ, Manguso RT, Cheruiyot CK, Bi K, Panda A, Iracheta-Vellve A, et al. Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade. Nature. 2019;565:43–8.
Dorhoi A, Du Plessis N. Monocytic myeloid-derived suppressor cells in chronic infections. Front Immunol. 2017;8:1895.
Acknowledgements
Immunofluorescence images were taken and flow cytometry data were analyzed at the Advanced Medical Research Institute, Shandong University. The authors thank Professor Haizhen Zhu (Hunan University) for the gift of the HLCZ-01 cell line. This work was supported by grants from the National Science Foundation of China (Key program 81830017, Nos. 81672425 and 81902051), the National Natural Science Fund for Outstanding Youth Fund (81425012), Taishan Scholarship (No. tspd20181201), Collaborative Innovation Center of Technology and Equipment for Biological Diagnosis and Therapy in Universities of Shandong, Key Research and Development Program of Shandong (2019GSF108238), the National Key Research and Development Program (2018YFE0126500 and 2016YFE0127000), China Mobility Grant jointly funded by the National Science Foundation of China and the Swedish Foundation for International Cooperation in Research and Higher Education (STINT), and China Postdoctoral Science Foundation (No. 2018 M30782).
Author information
Authors and Affiliations
Contributions
LW and ZCW carried out most of the experiments and analyzed data. YS contributed to the establishment of the protocols for the RIP and RNA pull-down assays. YS, ZHW, YZ, XP, XZ, and CL participated in the in vivo experiments. YKZ was involved with the IHC assay. CG provided help in the IFN pathway analysis. XL, NL, and LG were involved in the study design and manuscript preparation. LW wrote the manuscript with the help of CM. Author CM was in charge of the study design, work organization/supervision, and manuscript review. All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
About this article
Cite this article
Wang, L., Sun, Y., Song, X. et al. Hepatitis B virus evades immune recognition via RNA adenosine deaminase ADAR1-mediated viral RNA editing in hepatocytes. Cell Mol Immunol 18, 1871–1882 (2021). https://doi.org/10.1038/s41423-021-00729-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41423-021-00729-1
Keywords
This article is cited by
-
RNA editing enzymes: structure, biological functions and applications
Cell & Bioscience (2024)
-
RNA modification: mechanisms and therapeutic targets
Molecular Biomedicine (2023)
-
ADAR1p110 promotes Enterovirus D68 replication through its deaminase domain and inhibition of PKR pathway
Virology Journal (2022)
-
Molecular biology of autoinflammatory diseases
Inflammation and Regeneration (2021)