Abstract
Steroid receptor RNA activator (SRA)-like non-coding RNA (SLNCR1) has been implicated in various tumorigenic processes, but the precise regulatory role in melanoma progression remains uncertain. We performed a comprehensive analysis to investigate the prognostic value of SLNCR1 expression in patients with melanoma by TCGA database and melanoma tissue samples via the Kaplan–Meier method. Subsequently, we conducted qRT-PCR and Fluorescence in Situ Hybridization (FISH) assays to identify SLNCR1 expression levels and localization in tissues and cells, respectively. Loss-of-function assays utilizing shRNAs vectors were used to investigate the potential impact of SLNCR1. Our data showed that SLNCR1 is significantly up-regulated in human malignant melanoma tissues and cell lines and functions as an oncogene. Silencing of SLNCR1 suppressed melanoma cell proliferation, migration, invasion, and inhibited tumorigenesis in a mouse xenograft model. Additionally, we employed bioinformatic predictive analysis, combined with dual-luciferase reporter analysis and functional rescue assays, to elucidate the mechanistic target of the SLNCR1/SOX5 axis in melanoma. Mechanistically, we discovered that SLNCR1 promotes EMT of human melanoma by targeting SOX5, as downregulation of SLNCR1 expression leads to a decrease in SOX5 protein levels and inhibits melanoma tumorigenesis. Our research offers promising insights for more precise diagnosis and treatment of human melanoma.

Similar content being viewed by others
Introduction
Melanocytes originate from the neural crest and migrate during embryonic development to various parts of the body, such as skin, uvea, meninges, and mucous membranes [1]. Their malignant transformation causes melanoma. Risk factors associated with melanoma include sun exposure [2], the number and size of melanocyte nevi [3], and a family history of the disease [4]. Melanoma is the most lethal form of skin cancer, and early detection is key to successful treatment [5, 6]. Approximately 75% of melanomas are highly metastatic, and the incidence has increased annually in recent years [7]. Therefore, the development of early diagnostic and prognostic markers for melanoma is important for improving human health.
Long non-coding RNA (lncRNA), a class of non-protein-coding transcripts, are longer than 200 nts [8]. Growing evidence revealed that lncRNAs function as oncogenes or tumor suppressors, which can participate in tumor cell proliferation, migration, invasion, and apoptosis, through transcriptional and post-transcriptional regulatory mechanisms [9, 10].
Steroid receptor RNA activator (SRA)-like non-coding RNA (SLNCR1) is a recently identified lncRNA located on human chromosome 17q24.3 [11]. Despite being a relatively novel lncRNA, several studies have suggested that SLNCR1 expression is upregulated in various types of cancers, such as non-small cell lung cancer [12], breast cancer [13], cervical cancer [14], and papillary thyroid carcinoma [15], and may contribute to tumor progression. In a recent study, SLNCR1 was found to be involved in regulating melanoma invasion, but its specific functions and underlying mechanisms in melanoma remain unclear and merit further investigation [16].
This study focuses on exploring the role and mechanism of SLNCR1 and its downstream target gene in regulating the development of malignant melanoma. The transcription factor SRY-related high-mobility-group box5 (SOX5), a member of the SOX family, plays an important role in the regulation of gene transcription. SOX5 is essential for embryonic development and differentiation [17], and recent studies have highlighted its involvement in regulating proliferation, invasion, migration, metastasis, and epithelial-to-mesenchymal transition (EMT) in a variety of cancers [18,19,20,21]. Moreover, SOX5 has been shown to be associated with regulation by microphthalmia transcription factor (MITF) in melanoma [22].
In this study, we used the public database TCGA and melanoma tissue samples to clarify the expression and prognostic value of SLNCR1. Additionally, we assessed the functions of SLNCR1 both in vitro and in vivo using cell lines and mouse models. Finally, we explored the potential regulatory mechanisms by which SLNCR1 is involved in the development of melanoma progression. Our findings are expected to contribute new strategies for early diagnosis and precision treatment of melanoma.
Results
SLNCR1 is overexpressed and has a poor prognosis in malignant melanoma tissues and cell lines
To investigate the relationship between SLNCR1 expression and overall survival (OS), Kaplan–Meier analysis was performed using the TCGA database. Melanoma tissues were stratified into low or high groups based on the median expression of SLNCR1. Patients with high expression had worse OS than the low expression group (p = 0.027) (Fig. 1A). Additionally, 27 pairs of melanoma and adjacent tissues were collected and performed by qRT-PCR. SLNCR1 expression was significantly upregulated in melanoma tissues compared to adjacent tissue (Fig. 1B). This finding was further validated using various melanoma cell lines, including A375, A875, Mewo, Sk-mel-256, Sk-mel-888, and human immortalized keratinocytes (Hacat). The results demonstrated that the SLNCR1 expression was significantly increased in A375, A875, and Sk-mel-256 cell lines compared to Hacat cells (Fig. 1C). We have collected clinicopathology characteristics from 27 malignant melanoma patients in our hospital, the correlation between SLNCR1 expression and clinicopathologic characteristics of melanoma patients was also analyzed. The results showed that patients with high SLNCR1 expression may have a higher risk of P53 mutation (p = 0.0335) and lymph node metastasis (p = 0.0377) than patients with low expression (Table 1). SLNCR1 may act as an oncogene and be involved in melanoma progression.
A Kaplan–Meier OS curves of SLNCR1 expression from TCGA database. B SLNCR1 expression was determined using qPCR in 27 paired melanoma tissues and adjacent normal tissues. C Expression of SLNCR1 was assessed using qPCR in melanoma cell lines and normal cell lines. *p < 0.05; **p < 0.05; ***p < 0.005; N Normal, T Tumor.
SLNCR1 promotes melanoma cell proliferation, migration, invasion, and EMT
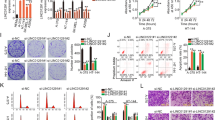
To identify how SLNCR1 expression regulates melanoma progression, we first localized SLNCR1 in the studied cell lines using RNA FISH assay. The results suggested that SLNCR1 was mainly located in the nucleus and a small amount distributed in the cytoplasm, indicating that SLNCR1 may promote melanoma progression at the level of transcriptional regulation (Fig. 2A). To determine the function of SLNCR1 in melanoma, loss-of-function assays were performed in A375 and A875 cells, respectively. As shown in Fig. 2B–D, SLNCR1 expression was significantly decreased after sh-SLNCR1 transfection which inhibited melanoma cell proliferation and colony formation. Additionally, the regulatory effect of SLNCR1 on cell metastasis was examined via wound healing and Transwell assays. Compared with the control group, the migration and invasion abilities were significantly reduced after SLNCR1 inhibition (Fig. 2E, F). Taken together, these results suggest that high SLNCR1 expression promotes melanoma proliferation, migration, and invasion.
A The FISH assay revealed the distribution of SLNCR1 in A375 and A875 cell lines. B Expression of SLNCR1 in the sh-SLNCR-transfected A375 and A875 cells was determined via qPCR. C, D CCK-8 and colony formation assays were conducted to determine the cell proliferation rate upon SLNCR1 silencing. E The effect of SLNCR1 interference on cell migration was estimated via wound healing assay. F Invasiveness of sh-SLNCR1- transfected A375 and A875 cell lines was evaluated via Transwell assay.
Epithelial-mesenchymal transition (EMT) with cell invasion, we investigated the changes in EMT markers after SLNCR1 knockdown to determine whether SLNCR1 was related to melanoma EMT. With SLNCR1 inhibition, both mRNA and protein levels of epithelial markers increased, while those of mesenchymal markers decreased, suggesting that SLNCR1 may promote EMT in melanoma (Fig. 3A, B).
SOX5 is the downstream target molecule of SLNCR1
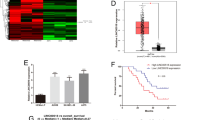
To investigate differentially expressed genes associated with SLNCR1 in melanoma progression, a |logFC| > 0.3 and p < 0.05 were considered the threshold for the SLNCR1-related genes by TCGA data using the R package (version 3.6.1). A total of 9889 upregulated and 3457 downregulated genes were screened (Fig. 4A). The top 20 differentially regulated candidate genes are summarized in Table 2.
A Volcano plot showed the differentially expressed transcriptional factors after SLNCR1 was knocked down. B Expression levels of the positively and negatively regulated molecules were detected after SLNCR1 was knocked down via qPCR. C Correlation between SLNCR1 and SOX5 expression level. D Kaplan–Meier overall survival of SOX5 in melanoma from TCGA database. E Expression levels of the genes in melanoma tissue predicted by GEPIA.
To validate the results of the analysis, we selected seven differentially expressed genes (four upregulated and three downregulated) and measured their expression levels in the SLNCR1-downregulated cell lines via qRT-PCR. As shown in Fig. 4B, among the positively correlated genes in both A375 and A875 cells, SOX5 expression was most significantly decreased after SLNCR1 knockdown. Based on TCGA data, we found that SLNCR1 expression was positively associated with SOX5 expression in melanoma (R = 0.54, p < 0.01) (Fig. 4C). Meanwhile, we noticed that high SOX5 expression was associated with shorter OS (p = 0.015) (Fig. 4D). Furthermore, results of GEPIA analysis showed that ACTB, SLC1A5, VASP, BRBB3, GAS7, ARTN2, and SOX5 were significantly increased in melanoma tissues (p < 0.05) (Fig. 4E). Therefore, SOX5 was selected as a candidate downstream target molecule of SLNCR1.
SOX5 is highly expressed in melanoma and regulates EMT
We next investigate the relationship between SOX5 expression and clinicopathologic features of patients. As shown in Table 3, SOX5 expression was significantly associated with N stage (p = 0.016), age (p = 0.046), tumor tissue site (p = 0.005), Breslow depth (p = 0.021), DSS event (p = 0.04), and PFI event (p = 0.004), indicating that SOX5 may participate in the occurrence and development of melanoma.
To further investigate the role of SOX5 in melanoma, we measured SOX5 expression levels in the 27 pairs of melanoma tissues and cells. As shown in Fig. 5A, SOX5 expression was significantly increased in melanoma tissues compared to normal tissues. Similar results were found in the cell lines, where both mRNA and protein expression of SOX5 were greatly increased in A375 and A875 cells (Fig. 5B, C). To determine the biological function of SOX5, siRNAs were transfected into these two melanoma cell lines (Fig. 5D). We found a significantly inhibitory effect on proliferation in the SOX5 silencing group compared to the control (Fig. 5E).
A SOX5 expression levels were determined via qPCR in the 27 paired melanoma tissues and adjacent normal tissues. B, C Both mRNA and protein expressions of SOX5 were measured via qPCR in the melanoma cell lines and normal cell lines. D Effects of SOX5 knockdown was determined using qPCR in A375 and A875 cell lines. E Effects of SOX5 inhibition on cell proliferation using CCK8 assay. Migration (F) and invasion (G) in A375 and A875 cells after SOX5 inhibition were observed by wound healing and transwell assays. mRNA expression (H) and protein expressions of EMT markers (I) in A375 and A875 cell lines after SOX5 inhibition were observed by qPCR and western blot assay.
Furthermore, to investigate migration and invasion in melanoma cells, wound healing and transwell assays were performed. As shown in Fig. 5F, G, compared to the control group, SOX5 inhibition significantly suppressed cell migration in both A375 and A875 cell lines. To further clarify the regulatory role of SOX5 on EMT, qPCR and western blot assays were performed. As shown in Fig. 5H, I, SOX5 inhibition resulted in a significant increase of E-cadherin, and a decrease in mesenchymal markers, including N-cadherin and vimentin. These results indicate that SOX5 may function as an oncogene involved in melanoma development.
SOX5 over-expression reverses si-SLNCR1 induced proliferation, migration, and EMT in vitro
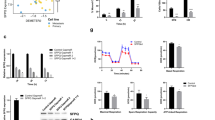
To investigate the potential interactions between SLNCR1 and SOX5, a dual-Luciferase reporter assay was performed. The results showed that silencing of SLNCR1 inhibited the transcriptional activity of the SOX5 promoter (Fig. 6A). Furthermore, SOX5 expression was significantly decreased after SLNCR1 knockdown in melanoma cells (Fig. 6B), indicating that SOX5 may be a downstream target molecule of SLNCR1. We then overexpressed SOX5 in SLNCR1-silenced melanoma cells to determine its effects on cell proliferation, migration, and EMT markers (Fig. 6C). As shown in Fig. 6D, E, SOX5 overexpression reversed the inhibition of cell proliferation and migration induced by SLNCR1 silencing. In addition, SOX5 overexpression had the opposite effect on the changes in EMT markers induced by SLNCR1 silencing (Fig. 6F, G). These findings suggest that SLNCR1 may regulate melanoma progression by targeting SOX5.
A SLNCR1 regulates the transcriptional activity at the SOX5 promoter region. B SOX5 expression was detected via qPCR after SLNCR1 was knocked down. C Overexpression of SOX5 was detected using qPCR. D CCK8 assay showed the proliferation of A375 and A875 cell lines when SOX5 was overexpressed. E Migration rates of A375 and A875 cells after SOX5 overexpressed. mRNA (F) and protein (G) expression of EMT markers after SOX5 overexpressed were observed by qPCR and western blot assays.
SLNCR1 down-regulation suppresses melanoma growth in vivo
To investigate the anti-tumor effects of SLNCR1 inhibition, we inoculated Balb/c mice with A375 cells stably expressing lentiviral sh-SLNCR1 to suppress SLNCR1 expression. Mice were randomly divided into two groups, one bearing sh-SLNCR1 cells and the other bearing sh-NC cells. After the treatment, the body weights and tumors were measured. As shown in Fig. 7A, compared to the normal group, significant tumor inhibitory effects were observed in the SLNCR1 inhibition group. After 2 weeks, the SLNCR1 inhibition group showed significantly decreased volumes and weights compared to the control (Fig. 7B, C). Enlargement and mitotic figures of nuclei were observed using H&E staining, indicating that subcutaneous tissue was tumor tissue (Fig. 7D). We also examined the heart, liver, spleen, lungs, and kidneys using histology. As shown in Fig. 7E, the knockdown of SLNCR1 had no obvious effect on the tissues of the mice internal organs. These results suggest that SLNCR1 could serve as a potential candidate target for melanoma treatment.
SLNCR1/SOX5 promotes melanoma by regulating EMT mechanism
In this study, we employed qPCR to investigate the in vivo relationship between SLNCR1 and SOX5. Our results revealed that SOX5 expression was significantly reduced in the sh-SLNCR1 group compared to the control group, indicating that SLNCR1 inhibition led to the down-regulation of SOX5 (Fig. 8A). To further validate this finding, we conducted an immunohistochemical (IHC) analysis to measure the protein expression of SOX5. The results were consistent with the qPCR findings, indicating that SLNCR1 inhibition resulted in decreased SOX5 expression (Fig. 8B). We also evaluated the expression of two epithelial-mesenchymal transitions (EMT) markers, Ki67 and Vimentin. Our findings showed that the knockdown of SLNCR1 resulted in the suppression of Ki67 and Vimentin expressions (Fig. 8C, D), suggesting that SLNCR1/SOX5 axis may promote melanoma growth in vivo via the regulation of the EMT pathway.
Discussion
Melanoma is a highly metastatic skin cancer [23]. Understanding the molecular pathways that govern the invasion of nevus melanocytes and the transformation of these cells into melanoma is crucial for fully elucidating the mechanisms underlying melanoma genesis. Many studies have shown that lncRNAs regulate various cellular processes, including cell proliferation, differentiation, migration, and invasion. Several lncRNAs, such as HOTAIR, MALAT1, and BANCR have been reported to be dysregulated in melanoma [24].
SLNCR1 is a newly discovered lncRNA that may play an essential role in the occurrence and development of melanoma. For example, SLNCR1 has been shown to promote melanoma invasion and growth through its interaction with androgen receptor and EGR1 [16, 25]. In our study, we further observed the function and role of SLNCR1 in malignant melanoma. Our findings revealed that SLNCR1 was overexpressed in melanoma tissues and cells, and analysis of TCGA database showed that high SLNCR1 expression was associated with poor overall survival rates. In this study, compared to Hacat cells, SLNCR1 is highly expressed in most of malignant melanoma cell lines, such as A375, A875, and sk-mel-256 cells. While Mewo and Sk- mel-888 cell lines obtained opposite results (Fig. 1C). This may be due to the different genetic backgrounds of different cell lines, including donor origin, mutation sites, and other species specificities. These results suggest that we could classify malignant melanoma into different subtypes based on SLNCR1 expression for precision diagnosis in the future.
Furthermore, knockdown of SLNCR1 inhibited the proliferation, migration, and invasion of melanoma cells in vitro. Since cell invasion is closely related to EMT [26], we evaluated EMT markers following SLNCR1 knockdown. We found that the expression of epithelial marker E-cadherin was up-regulated, while the mesenchymal markers N-cadherin and vimentin were down-regulated, suggesting that SLNCR1 knockdown suppressed the occurrence of EMT.
Different localization of lncRNA in cells determines how they participate in tumor progression [27]. The lncRNA in the nucleus usually participates in melanoma progression in the transcriptional regulation way by modulating their downstream transcription factors [28]. It was found that lncRNA BASP1-AS1 can interact with YBX1, activating the Notch signaling pathway and driving migration in melanoma [29]. The LncRNA in the cytoplasm is involved in cancer in the post-transcriptional regulation way by interacting with specific proteins and RNAs [30]. LncRNA HOXD-AS1 can bind to EZH2 and inhibit RUNX3 expression through epigenetic regulation [31]. Furthermore, there is considerable evidence that competition for miRNAs plays an important part in LncRNA regulation. For example, lncRNA MALAT1 could bind to miR-23a, promoting proliferation, migration, and invasion of melanoma [32]. Although diverse functions have been characterized, specific insights relating to the lncRNAs in tumorigenesis are poorly understood.
SLNCR1 was mainly localized in the nucleus of cells using RNA FISH. Therefore, we inferred that SLNCR1 promotes melanoma genesis through transcriptional regulation. To identify the downstream transcription factors of SLNCR1, we searched the TCGA database and found that SOX5 may be a target molecule of SLNCR1. SOX5 plays an essential role in cell-fate decision and differentiation. For example, SOX5 could inhibit glioma proliferation in vitro, while SOX5 knockdown elevates the ability of glioma growth in mouse models [33]. Moreover, SOX5 has been identified as a predictor of poor prognosis in lung adenocarcinoma, and SOX5 is known to promote lung adenocarcinoma progression and metastasis through EMT [34].
In our study, GEPIA predicted that SOX5 was overexpressed; therefore, qPCR and western blot analyses were performed. SOX5 was highly expressed in melanoma tissues and cell lines compared to controls. Furthermore, SOX5 silencing suppressed the proliferation and migration capability of melanoma cells, but not invasion. We found that the expression of SOX5 decreased after SLNCR1 knockdown, and dual-luciferase reporter assays showed that SLNCR1 silencing reduced the transcriptional activity of the SOX5 promoter region, which verified the regulatory relationship between SLNCR1 and SOX5. Overexpression of SOX5 reversed the inhibition of proliferation and migration of melanoma cells induced by SLNCR1 silencing.
SLNCR1, a novel lncRNA with potential cancer-promoting function, which downstream regulated target genes and functions are still unclear. In our study, we found downstream target SOX5 of SLNCR1 from the perspective of lncRNA transcriptional regulation and clarified that SLNCR1/SOX5 axis promotes invasion and metastasis of melanoma through EMT, suggesting that SLNCR1 could be used as a potential biomarker and therapeutic target for melanoma.
This study successfully constructed a subcutaneous tumorigenesis model in nude mice using SLNCR1-stable knockdown A375 cells. Our results showed that SLNCR1 silencing inhibited melanoma proliferation and EMT in vitro. Furthermore, SLNCR1 inhibition decreased both mRNA and protein expression of SOX5, which were consistent with the previous results.
In conclusion, our study revealed that SLNCR1 promotes EMT of melanoma by targeting SOX5, these findings suggest that SLNCR1 may serve as a potential marker for assessing the incidence and prognosis of melanoma.
Materials and methods
Data collection and prognostic model
RNA-seq transcriptomic data and clinical data for 472 melanoma patients were downloaded from the Cancer Genome Atlas (TCGA) database (https://portal.gdc.caner.gov). Of these patients, 449 had complete survival data, and we classified them into high- and low-risk groups based on their risk scores, with the median value serving as the threshold. Kaplan–Meier method was performed for overall survival (OS).
We retrieved gene expression data for skin malignant melanoma from the official TCGA database using GDC-client and obtained the original RNA expression data. To standardize the data, we converted it to TPM format, resulting in a total of 17,580 genes, including SLNCR1, after filtering out those with very low expression values. We then performed log2 transformation on these genes and conducted a correlation analysis to investigate the relationship between SLNCR1 and other coding genes.
GEPIA analysis
GEPIA (http://gepia.cancer-pku.cn/index.html) is an open-access database that enables in-depth analysis of gene expression data from TCGA. GEPIA was performed to investigate candidate genes associated with SLNCR1.
Patients and samples
A total of 27 pairs of melanoma and adjacent tissues were collected from China-Japan Union Hospital of Jilin University (2009–2021). These patients were diagnosed with melanoma through pathological examination and had not undergone any chemotherapy or radiotherapy before surgical resection. The adjacent tissues were defined as having no tumor at the surgical margin. Table 1 provides a summary of the clinicopathological characteristics of the patients included in this study. Ethical approval was obtained by the China-Japan Union Hospital of Jilin University.
Cell culture
The human malignant melanoma cell lines A375 and A875 were purchased from ATCC, while the Hacat, Mewo, Sk-mel-256, and Sk-mel-888 cell lines were maintained in our laboratory. All cell lines were cultured in DMEM with 10% fetal bovine serum.
Silencing of target gene expression by siRNA and shRNA
Small interfering RNA (siRNA) targeting SOX5 and control siRNA were designed by Genepharma (Shanghai, China). According to the manufacturer’s instructions, cell transfections were performed using Lipofectamine 2000 (Invitrogen, USA). qPCR analysis to assess the efficacy of knockdown. We purchased the SLNCR1 lentiviral vector and empty lentiviral vector from Hanheng (Shanghai, China), infected cells and selected with 1 μg/ml puromycin (Cat#P8230, Solarbio, China) to obtain stable knockdown cell lines.
RNA extraction and real-time quantitative PCR (qPCR)
Total RNA was extracted using Trizol Reagent (Invitrogen) following the manufacturer’s instructions. cDNA was reverse transcripted using an RR047A kit (Takara, Japan). Real-time PCR was performed with SYBR Green (Roche, USA) on a 7500 Fast Real-Time PCR system (ABI, USA). Gene expression was quantified using the 2−ΔΔCt method, with actin expression used as the internal control for normalization. The primers used are listed in Table 4.
Cell counting kit-8 (CCK8) assay
To assess the effect of SLNCR1 on cell proliferation, cell Counting Kit-8 (CCK-8) assay was performed. Cells were seeded in 96-well plates at a density of 2 × 10^3 cells/well. CCK-8 solution was added and incubated for 2 h. The absorbance was observed by a microplate at 450 nm (Bio-tek).
Wound healing assay
To evaluate the effect of SLNCR1 on melanoma cell migration, a wound healing assay was performed Approximately 5 × 10^4 cells were seeded in a 6-well plate. After 48 h, the cells were wounded to create a straight scratch. The wells were then washed and treated with fresh medium containing 2% FBS. Digital microscopy (Olympus) was used to capture images of each wound at 0, 24, and 48 h. The wound space was calculated by subtracting the scratch widths at 24 or 48 h from the width at 0 h using the MRI tool in Image J.
Transwell invasion assay
To determine the invasive abilities, Transwell invasion assay was performed. A total of 5 × 10^3 cells without serum were added to the upper chamber of Transwell inserts (Corning) coated with matrigel (BD Biosciences). DMEM medium containing 10% FBS was added to the lower chamber as a chemoattractant. After 48 h, the invaded or migrated cells were fixed and stained. Non-invading cells were removed. The invasive abilities were evaluated in five randomly selected fields.
Western blot assay
The cells were harvested and lysed, measured using bicinchoninic acid (BCA) protein assay kits (Beyotime). Then, the samples were subjected to SDS-PAGE and transferred. The blot was incubated with primary antibodies against E-cadherin (1:1000, cat#198751, Sangon), N-cadherin (1:1000, cat#D199282, Sangon), vimentin (1:1000, cat#5741, CST), Slug (1:1000, cat#9585, CST), SOX5 (1:1000, cat#13216-1-AP, Proteintech), and GAPDH. The bands were visualized using a chemiluminescence imaging system (SAGE).
Luciferase reporter assay
In the initial step, a dual-luciferase reporter plasmid, pGL3-basic, containing the promoter region (1-600) of SOX5 was synthesized by Comate Biosciences (pGL3- basic-SOX5-promoter). Cells were seeded at 5 × 10^4 cells/well in 24-well plates and allowed to grow overnight. Then, the cells were co-transfected with pGL3-Basic- SOX5-promoter, SLNCR1-siRNA, and Renilla plasmids (Comate Biosciences, Shanghai, China). After transfection, the cells were lysed and the supernatant was collected. Firefly luciferase reagent was added, and the luciferase activity was measured using a Multilabel Reader (PerkinElmer) and normalized to the renilla luciferase activity.
Fluorescence in situ hybridization (FISH)
The FISH probe was synthesized by Genepharma. According to the manufacturer’s instructions, cells were seeded onto confocal plates and incubated overnight. Then, the cells were washed with PBS and fixed. The cells were then treated with protease K, glycine, and acetylation reagent, incubated with hybridization solution, and probe-labeled SLNCR1. The cover glass was washed with PBST and the nucleus was stained with DAPI (1:800) diluted in PBST. The confocal plates were washed, sealed, and observed by fluorescence microscope (Olympus).
Hematoxylin-eosin (H&E) staining
The organs were fixed overnight. Subsequently, the tissues were stained with hematoxylin and eosin (H&E) and observed by microscope (Olympus, Tokyo, Japan).
Immunohistochemistry (IHC)
Immunohistochemical (IHC) staining was performed on dissected subcutaneous tumors from mice. The slides were deparaffinized and dehydrated with xylene and graded alcohols, respectively. Antigen retrieval was performed, and the slides were incubated with primary antibodies against SOX5, vimentin, and Ki67, followed by incubation with a secondary antibody (DAKO) and visualization using DAB solution.
Xenograft model assay
A375 melanoma cells stably transduced with either sh-NC (control) or sh-SLNCR1 were cultured in complete media supplemented with 1 μg/ml puromycin until reaching confluence. Ten 6-week-old female BALB/c nude mice were subcutaneously injected with A375-NC or A375-sh-SLNCR1 transduced cells at a dose of 1 × 10^7/100 μL into the right back. Tumor size was measured on days 6, 8, 10, 12, and 14 after injection, and the animals were euthanized when the tumor volumes reached about 100 mm3. Tumor volume was calculated using the formula V(mm3) = (L × W2) × 0.5 (L: tumor length, W: width). All the animal experiment was approved by Institional Animal Ethics Committee.
Statistic analysis
Statistical analysis was performed using GraphPad Prism 8.0 software (GraphPad Software, Inc.). Student’s t test was used to compare the means of two independent samples. The associations between SLNCR1 mRNA levels and clinical pathological parameters of melanoma patients were assessed using the Chi-square (χ2) test. Survival curve analyses were performed using the R package (version 3.6.1). The expression of SOX5 was estimated using the GEPIA and TCGA databases. Statistical significance was defined as follows: *p < 0.05; **p < 0.005; and ***p < 0.0005.
Data availability
All data generated or analyzed during this study are included in this published article.
References
Strashilov S, Yordanov A. Aetiology and Pathogenesis of Cutaneous Melanoma: Current Concepts and Advances. Int J Mol Sci. 2021;22:6395.
Gandini S, Sera F, Cattaruzza MS, Pasquini P, Picconi O, Boyle P, et al. Meta-analysis of risk factors for cutaneous melanoma: II. Sun exposure. Eur J Cancer. 2005;41:45–60.
Shain AH, Bastian BC. From melanocytes to melanomas. Nat Rev Cancer. 2016;16:345–58.
Soura E, Eliades PJ, Shannon K, Stratigos AJ, Tsao H. Hereditary melanoma: Update on syndromes and management: Genetics of familial atypical multiple mole melanoma syndrome. J Am Acad Dermatol. 2016;74:395–407.
Swetter SM, Tsao H, Bichakjian CK, Curiel-Lewandrowski C, Elder DE, Gershenwald JE, et al. Guidelines of care for the management of primary cutaneous melanoma. J Am Acad Dermatol. 2019;80:208–50.
Han W, Shen GL. Systematic expression analysis of EAF family reveals the importance of EAF2 in melanoma. Int Immunopharmacol. 2020;88:106958.
Ali Z, Yousaf N, Larkin J. Melanoma epidemiology, biology and prognosis. EJC Suppl. 2013;11:81–91.
Nair L, Chung H, Basu U. Regulation of long non-coding RNAs and genome dynamics by the RNA surveillance machinery. Nat Rev Mol Cell Biol. 2020;21:123–36.
Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145:178–81.
Yarmishyn AA, Kurochkin IV. Long noncoding RNAs: a potential novel class of cancer biomarkers. Front Genet. 2015;6:145.
Cong L, Sun H, Hao M, Sun Q, Zheng Y, Cong X, et al. The Prognostic Value of LncRNA SLNCR1 in Cancers: A Meta-Analysis. J Oncol. 2021;2021:3161714.
Xu W, Xu Q, Kuang D, Wang Z, Lu Q, Lin Q, et al. Long non‑coding RNA SLNCR1 regulates non‑small cell lung cancer migration, invasion and stemness through interactions with secretory phospholipase A2. Mol Med Rep. 2019;20:2591–6.
Qiao K, Ning S, Wan L, Wu H, Wang Q, Zhang X, et al. LINC00673 is activated by YY1 and promotes the proliferation of breast cancer cells via the miR-515-5p/MARK4/Hippo signaling pathway. J Exp Clin Cancer Res. 2019;38:418.
Shi WJ, Liu H, Ge YF, Wu D, Tan YJ, Shen YC, et al. LINC00673 exerts oncogenic function in cervical cancer by negatively regulating miR-126-5p expression and activates PTEN/PI3K/AKT signaling pathway. Cytokine. 2020;136:155286.
Meng XF, Zhao LY, Chu XF. LncRNA LINC00673 inhibits p53 expression by interacting with EZH2 and DNMT1 in papillary thyroid carcinoma. Eur Rev Med Pharmacol Sci. 2019;23:2075–83.
Schmidt K, Joyce CE, Buquicchio F, Brown A, Ritz J, Distel RJ, et al. The lncRNA SLNCR1 Mediates Melanoma Invasion through a Conserved SRA1-like Region. Cell Rep. 2016;15:2025–37.
Baroti T, Zimmermann Y, Schillinger A, Liu L, Lommes P, Wegner M, et al. Transcription factors Sox5 and Sox6 exert direct and indirect influences on oligodendroglial migration in spinal cord and forebrain. Glia. 2016;64:122–38.
Sun C, Ban Y, Wang K, Sun Y, Zhao Z. SOX5 promotes breast cancer proliferation and invasion by transactivation of EZH2. Oncol Lett. 2019;17:2754–62.
Renjie W, Haiqian L. MiR-132, miR-15a and miR-16 synergistically inhibit pituitary tumor cell proliferation, invasion and migration by targeting Sox5. Cancer Lett. 2015;356:568–78.
Grimm D, Bauer J, Wise P, Krüger M, Simonsen U, Wehland M, et al. The role of SOX family members in solid tumours and metastasis. Semin Cancer Biol. 2020;67:122–53.
Hu J, Tian J, Zhu S, Sun L, Yu J, Tian H, et al. Sox5 contributes to prostate cancer metastasis and is a master regulator of TGF-β-induced epithelial mesenchymal transition through controlling Twist1 expression. Br J Cancer. 2018;118:88–97.
Aguennouz M, Guarneri F, Oteri R, Polito F, Giuffrida R, Cannavò SP. Serum levels of miRNA-21-5p in vitiligo patients and effects of miRNA-21-5p on SOX5, beta-catenin, CDK2 and MITF protein expression in normal human melanocytes. J Dermatol Sci. 2021;101:22–29.
Dorrell DN, Strowd LC. Skin Cancer Detection Technology. Dermatol Clin. 2019;37:527–36.
Yu X, Zheng H, Tse G, Chan MT, Wu WK. Long non-coding RNAs in melanoma. Cell Prolif. 2018;51:e12457.
Schmidt K, Carroll JS, Yee E, Thomas DD, Wert-Lamas L, Neier SC, et al. The lncRNA SLNCR Recruits the Androgen Receptor to EGR1-Bound Genes in Melanoma and Inhibits Expression of Tumor Suppressor p21. Cell Rep. 2019;27:2493–507.
Pastushenko I, Blanpain C. EMT Transition States during Tumor Progression and Metastasis. Trends Cell Biol. 2019;29:212–26.
Bridges MC, Daulagala AC, Kourtidis A. LNCcation: lncRNA localization and function. J Cell Biol. 2021;220:e202009045.
Xing C, Sun SG, Yue ZQ, Bai F. Role of lncRNA LUCAT1 in cancer. Biomed Pharmacother. 2021;134:111158.
Li Y, Gao Y, Niu X, Tang M, Li J, Song B, et al. LncRNA BASP1-AS1 interacts with YBX1 to regulate Notch transcription and drives the malignancy of melanoma. Cancer Sci. 2021;112:4526–42.
Dykes IM, Emanueli C. Transcriptional and Post-transcriptional Gene Regulation by Long Non-coding RNA. Genomics Proteomics Bioinformatics. 2017;15:177–86.
Zhang H, Bai M, Zeng A, Si L, Yu N, Wang X. LncRNA HOXD-AS1 promotes melanoma cell proliferation and invasion by suppressing RUNX3 expression. Am J Cancer Res. 2017;7:2526–35.
Wang P, Hu L, Fu G, Lu J, Zheng Y, Li Y, et al. LncRNA MALAT1 Promotes the Proliferation, Migration, and Invasion of Melanoma Cells by Downregulating miR-23a. Cancer Manag Res. 2020;12:6553–62.
Kurtsdotter I, Topcic D, Karlén A, Singla B, Hagey DW, Bergsland M, et al. SOX5/6/21 Prevent Oncogene-Driven Transformation of Brain Stem Cells. Cancer Res. 2017;77:4985–97.
Chen X, Fu Y, Xu H, Teng P, Xie Q, Zhang Y, et al. SOX5 predicts poor prognosis in lung adenocarcinoma and promotes tumor metastasis through epithelial-mesenchymal transition. Oncotarget. 2017;9:10891–904.
Funding
This research was supported by the Department of Science and Technology of Jilin Province (grant no. 20230402003GH, YDZJ202201ZYTS021), and Chunlei project of China-Japan Union Hospital of Jilin University (grant no. 2023CL07). The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
MH conceived the project. LC, QZ, HS, and YH designed experiments, and interpreted data in the manuscript. ZZ performed bioinformatics analyses. LC wrote the manuscript. MH, CL, and XC edited the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Collection of the melanoma tissues was approved by the Medical Ethics Committee of China-Japan Union Hospital of Jilin University, and written informed consent was obtained from each patient. Title of the approved project: Study on the pathogenesis of malignant melanoma. Approval number: 2019071606. Date of approval: July 16, 2019. All animal experimental procedures were approved by the Institutional Animal Care and Use Committee of Jilin University (Changchun, China). Title of the approved project: Effects of lncRNA SLNCR1 on malignant melanoma. Approval number: 20211109-01. Date of approval: November 5, 2021.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cong, L., Zhao, Q., Sun, H. et al. A novel long non-coding RNA SLNCR1 promotes proliferation, migration, and invasion of melanoma via transcriptionally regulating SOX5. Cell Death Discov. 10, 160 (2024). https://doi.org/10.1038/s41420-024-01922-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41420-024-01922-7