Abstract
The pluripotency of naïve mouse embryonic stem cells (mES) is regulated by multiple signaling pathways, with inhibition of protein kinase C (PKCi) playing a particularly important role in maintaining naïve mES. However, the regulatory function of nucleosome remodeling and deacetylase (NuRD) complex in mES cultured in a PKCi system is unknown. We found that, compared with 2iL-derived mES, PKCi-derived mES showed low mRNA expression of NuRD complex subunits, including MBD3, HDAC1/HDAC2, MTA1, and RbAP46/RbAP48. Western blot showed that PKCi-derived mES expressed lower protein levels of MBD3 and HDAC2 at passage 3, as well as MBD3, HDAC2, and MTA1 at passage 10, indicating that PKCi suppressed NuRD complex expression. Knockdown of MBD3 increased PKCi-derived mES pluripotency by increasing NANOG and OCT4 expression and colony formation. By contrast, overexpression of MBD3 or removal of PKC inhibitor-induced differentiation of mES, results in reduced NANOG, OCT4, and REX1 expression and colony formation, increased differentiation-related gene expression, and differentiation into flat cells. Knockdown of MBD3 in mES upon PKC inhibitor removal partially reversed cell differentiation. Our results show that the regulatory NuRD complex and its MBD3 subunit influence the naïve pluripotency of mES cultured in a PKCi system.
Similar content being viewed by others
Introduction
The nuclear remodeling and deacetylation (NuRD) complex is an abundant and conserved regulator of chromatin structure remodeling and transcriptional repression [1, 2]. The NuRD complex contains several subunits (e.g., methyl-CpG-binding domain protein MBD2/3, histone deacetylase core proteins HDAC1/2, metastasis-associated protein MTA1/2/3, ATP-dependent nucleosome remodeling enzyme CHD3/4, histone-binding/chaperone proteins RbAP46/48, zinc-finger proteins p66α/β, and DOC1), mediates two major biological functions: nucleosome remodeling in chromatin formation and histone deacetylation, resulting in the silencing of gene transcription [3]. NuRD was recently found to modulate chromatin structure at regulatory elements of active transcription sites, thereby regulating gene expression in a finely tuned manner [1].
Embryonic stem cells (ES), first derived by Evans and Kaufman in 1981 [4], possess the characteristics of self-renewal, indefinite proliferation in vitro, multi-lineage differentiation, and germline transmission. Mouse ES (mES) can exist in naïve or primed states [5]. Two different culture systems can maintain mES naïve pluripotency: 2iL and inhibition of the PKC signaling pathway (PKCi). The 2iL system includes leukemia inhibitory factor (LIF), which activates transcription factor signal transducers and activators of transcription 3 (STAT3) and the small-molecule inhibitors PD0325901 and CHIR99021, which in turn inhibit mitogen-activated protein kinase (MAPK) and glycogen synthase kinase 3 (GSK3) pathways, respectively [6, 7]. 2iL-derived mES are frequently used in stem cell studies. The PKCi system also maintains the naïve state of mES and rat ESs [8, 9]. Members of the PKC family are intracellular mediators of several hormones, neurotransmitters, phorbol esters, and tumor promoters that play essential roles in growth regulation, ES self-renewal, cell differentiation, neurotransmission, and cell death [10]. In particular, inhibition of the PKCζ–NF-κB–microRNA-21/microRNA-29 axis is key to maintaining ES self-renewal and naïve pluripotency [8, 9]. However, the role of NuRD subunits in PKCi-derived naïve mES, as well as differences in NuRD complex expression between 2iL- and PKCi-derived mES, are unknown.
Previous studies revealed the mechanisms by which NuRD maintains stem cell pluripotency via the regulation of gene expression related to cell plasticity, self-renewal, and differentiation during development [1, 11]. MBD3, a specific ES NuRD subunit, is essential for early embryogenesis and the development of pluripotent stem cells [12, 13]. As a methylated DNA binding protein that scaffolds other NuRD subunits, MBD3 is crucial for recruiting other subunits and assembling the NuRD complex [14, 15] and modulates transcriptional heterogeneity, and maintains ES lineage commitment by directly regulating the expression of pluripotency genes in ESs [16]. MBD3 is required for commitment to a full spectrum of embryonic lineages [15, 17, 18]. MBD3−/− ES maintains the expression of pluripotency genes such as OCT4 and NANOG but fails to form stable NuRD complexes, exhibit severe defects in differentiation [12], and shows silencing of Oct4 expression upon withdrawal of LIF [15]. However, the role of MBD3 in the growth and self-renewal of mES colonies in the PKCi system is unclear. In particular, identifying differences in target gene regulatory loci specifically recognized by MBD3 between 2iL- and PKCi-derived mES will provide a fuller understanding of how different signaling pathways induce naïve mES.
In this study, we examined differences in NuRD complex RNA and protein expression in naïve mES derived in 2iL versus PKCi culture systems. We found that NuRD subunits are present in both types of mES, suggesting that NuRD is necessary for maintaining ES self-renewal and pluripotency. However, the RNA expression of most NuRD subunits, including HDAC2, MBD3, and MTA1, was lower in PKCi-derived mES than in 2iL-derived mES. Furthermore, using overexpression and knockdown of MBD3, we demonstrate that NuRD and MBD3 play important roles in PKCi-derived mES self-renewal and naïve pluripotency by modulating the expression of pluripotency and differentiation genes.
Results
PKCi downregulated expression of the NuRD complex in mES
To investigate how the PKCi culture system affects the expression of NuRD complex components, we first measured the mRNA expression of the NuRD complex in PKCi-derived mES at passage 3. Quantitative polymerase chain reaction (qPCR) showed that compared with 2iL-derived mES, PKCi-derived mES showed lower mRNA levels of MBD3 (from 1.0 to 0.2), HDAC2 (from 1.0 to 0.4), HDAC1 (from 1.0 to 0.3), MTA1 (from 1.0 to 0.1), RbAP46 (from 1.0 to 0.02), RbAP48 (from 1.0 to 0.05), and p66α (from 1.0 to 0.8) (P < 0.05; Fig. 1A), whereas CHD3, p66β, and DOC1 levels were similar between two mES types. Western blot further revealed that PKCi-derived mES showed lower protein levels of MBD3 (from 1.0 to 0.5) and HDAC2 (from 1.0 to 0.9, P < 0.05; Fig. 1B). These changes in RNA and protein expression were maintained (Fig. 1C, D), except for further reduced protein MTA1 (Fig. 1D) at passage 10. When we examined the expression of genes downstream and targets of NuRD [1], we found significantly lower expression of carbonyl reductase 3 (CBR3) and high-temperature requirement serine protease A1 (HTRA1) mRNA in PKCi-derived mES (Fig. 1E).
A qPCR showed that compared with 2iL, PKCi decreased mRNA levels of NuRD complex subunits MBD3, HDAC2, HDAC1, MTA1, RbAP46, RbAP48, and p66α, but not of CHD3, p66β, and DOC1, in mES at passage 3. B Protein levels of NuRD complex and β-actin were evaluated by Western blot (left panel). Quantitative density analysis showed that compared with 2iL, PKCi decreased protein levels of MBD3 and HDAC2, but not other subunits (right panel). C qPCR showed that compared with 2iL, PKCi decreased mRNA levels of MBD3, HDAC2, HDAC1, MTA1, RbAP46, RbAP48, and p66α, but not of CHD3, p666β, and DOC1, in mES at passage 10. D Protein levels of NuRD complex and β-actin were evaluated by Western blot (left panel). Quantitative density analysis showed that compared with 2iL, PKCi decreased protein levels of MBD3, HDAC2, and further MTA1, but not of other subunits (right panel). E qPCR showed that compared with 2iL, PKCi significantly decreased the mRNA levels of NuRD target genes including CBR3 and HTRA1 in mES at passage 3. Data were shown as mean ± SD (n = 3). The letters a and b indicated significant differences among groups (P < 0.05).
PKCi-derived mES expressed naïve pluripotency genes and possessed germline transmission
Compared with mouse embryonic fibroblasts, PKCi- and 2iL-derived mES showed higher mRNA expression of the pluripotency markers such as NANOG, OCT4, C-MYC, and SOX2 (Fig. S1A) and naïve-state markers FGF4, NROB1, REX1, and KLF4 (Fig. S1B) but not primed-state markers FGF5 and T (P < 0.05; Fig. S1C).
Signaling pathways involved in PKCi-derived mES self-renewal
As WNT, ERK, and AKT signaling pathways are involved in the self-renewal of 2iL-derived mES [6, 7], we examined key proteins in these pathways in both 2iL- and PKCi-derived mES. Western blot showed that the ratio of phosphorylated (p)-β-catenin/β-catenin was increased in PKCi-derived mES (P < 0.01), whereas p-ERK/ERK and p-AKT/AKT ratios were similar between two mES types (Fig. 2A, B). In addition, levels of HDAC5, a key protein in the PKCµ signaling pathway [19], were reduced in PKCi-derived mES (P < 0.01; Fig. 2).
A p-β-catenin/β-catenin, HDAC5, p-ERK/ERK, p-AKT/AKT, and β-actin protein levels were assessed by Western blot in PKCi-derived mES at passage 3, with 2iL-derived mES used as a positive control. B Quantitative density analysis showed that PKCi increased protein levels of p-β-catenin/β-catenin and decreased the level of HDAC5. Data were shown as mean ± SD (n = 3). The letters a and b indicated significant differences among groups (P < 0.05).
MBD3 knockdown promoted PKCi-derived mES self-renewal
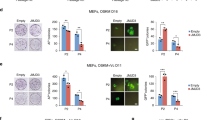
To understand the role of MBD3 in mES self-renewal, we knocked down MBD3 in PKCi-derived mES. Compared with PKCi and shNC control groups, MBD3 mRNA (from 1.0 to 0.3) (Fig. 3A) and protein (from 1.0 to 0.4) (Fig. 3B) expression were reduced in MBD3 knocked-down mES (P < 0.01). MBD3 knockdown increased the mRNA levels of pluripotency markers NANOG (from 1.0 to 3.7) and OCT4 (from 1.0 to 2.6) (P < 0.01), whereas the expression of other naïve pluripotency, primed-state, and differentiation genes was unchanged (Fig. 3C). As expected, Western blot revealed that the protein levels of NANOG (from 1.0 to 2.0) and OCT4 (from 1.0 to 3.0) were also increased (P < 0.01; Fig. 3D). Immunostaining indicated that MBD3 knockdown did not affect the morphology of mES (Supporting Information Fig. S1D). Although AP staining indicated that compared with PKCi and control groups, MBD3 knockdown did not affect the total number of AP-positive colonies, the percentage of mixed colonies increased (from 42.1% to 46.4%) and differentiated colonies decreased (from 9.1% to 3.8%), respectively (P < 0.05; Fig. 3E).
A qPCR showed that mRNA levels of MBD3 decreased after PKCi-derived mES at passage 5 were transfected with shMBD3, with PKCi and shNC groups used as controls. B MBD3 and β-actin protein levels were evaluated by Western blot in PKCi-derived mES with MBD3 knockdown (upper panel). Quantitative density analysis showed that shMBD3 decreased protein levels of MBD3 (lower panel). C qPCR showed that compared with the control groups, shMBD3 increased the mRNA levels of pluripotency genes NANOG and OCT4 (left panel) but did not affect the mRNA levels of differentiation genes (right panel). D Western blot detection of NANOG, OCT4, and β-actin protein expression in PKCi-derived mES with MBD3 knockdown (left panel). Quantitative density analysis showed that shMBD3 increased protein levels of NANOG and OCT4 (right panel). E Knockdown of MBD3 did not affect the morphology or AP staining of PKCi-derived mES. Scale bar, 200 μm (upper panel). Knockdown of MBD3 did not affect the total number of AP-positive colonies (left panel) but increased the percentage of mixed colonies and decreased the percentage of differentiated colonies (right panel). Data were shown as mean ± SD (n = 3). The letters a and b indicated significant differences among groups (P < 0.05).
Overexpression of MBD3-induced mES differentiation
Compared with PKCi and FUW-M2rtTA control groups, MBD3 overexpression increased mRNA (from 1.0 to 2.0; Fig. 4A) and protein (from 1.0 to 1.4, P < 0.05; Fig. 4B) levels of MBD3 in PKCi-derived mES. qPCR showed that MBD3 overexpression significantly decreased mRNA levels of the pluripotency markers NANOG (from 1.0 to 0.5) and OCT4 (from 1.0 to 0.8) and the naïve-state marker REX1 (from 1.0 to 0.2) and increased mRNAs levels of the primed-state marker FGF5 (from 1.0 to 4.2). mRNA levels of the differentiation markers CK8 (from 1.0 to 5.6), cTnT (from 1.0 to 3.6), BMP4 (from 1.0 to 1.9), DESMIN (from 1.0 to 1.4), PAX6 (from 1.0 to 4.4), and SOX17 (from 1.0 to 1.8) were increased in MBD3-overexpressing mES (P < 0.05; Fig. 4C). Also, MBD3 overexpression decreased protein levels of NANOG (from 1.0 to 0.3) and OCT4 (from 1.0 to 0.2) and increased protein levels of cTnT (from 1.0 to 3.0, P < 0.05; Fig. 4D).
A qPCR showed that mRNA levels of MBD3 increased after PKCi-derived mES at passage 5 were transfected with FUW-MBD3, with the PKCi and FUW-M2rtTA plasmid groups used as controls. B MBD3 and β-actin protein levels were evaluated by Western blot after PKCi-derived mES were transfected with FUW-MBD3 (upper panel). Quantitative density analysis showed that protein levels of MBD3 increased after transfection with FUW-MBD3 (lower panel). C qPCR showed decreased mRNA levels of pluripotency genes NANOG and OCT4 and naïve-state marker REX1, but increased mRNA levels of primed-state marker FGF5 after MBD3 was overexpressed (left panel). MBD3 overexpression also increased mRNA levels of the endoderm marker CK8, mesoderm markers cTnT, BMP4, and DESMIN, and ectoderm markers PAX6 and SOX17 (right panel). D Western blot detection of NANOG, OCT4, REX1, FGF5, cTnT, and β-actin protein expression after MBD3 overexpression (left panel). Quantitative density analysis showed that MBD3 overexpression decreased protein levels of NANOG, OCT4, and REX1, but increased protein levels of FGF5 and cTnT (right panel). E MBD3 overexpression induced PKCi-derived mES differentiation and resulted in a loss of AP staining. Scale bar, 200 μm (upper panel). Overexpression of MBD3 reduced the total number of AP-positive colonies (left panel), decreased the percentage of undifferentiated and mixed colonies, and increased the percentage of differentiated colonies (right panel). Data were shown as mean ± SD (n = 3). The letters a and b indicated significant differences among groups (P < 0.05).
MBD3 overexpression induced mES differentiation into flat cells that did not express NANOG (Supporting Information Fig. S2C). Compared with the PKCi control group, MBD3 overexpression reduced the total number of AP-positive colonies (from 178 to 50) and the percentage of undifferentiated (from 45.4% to 6.1%) and mixed (from 46.0% to 13.4%) colonies, but increased the percentage of differentiated colonies (from 8.7% to 80.5%, P < 0.05; Fig. 4E).
Removal of PKC inhibitor increased MBD3 expression and mES differentiation, which was partially rescued by MBD3 knockdown
When the PKC inhibitor Gӧ6983 was removed from the PKCi culture medium for 48 h, MBD3 mRNA (from 1.0 to 2.1) and protein (from 1.0 to 1.7) levels increased in mES (P < 0.05; Fig. 5A, B). This change was associated with decreased mRNA levels of pluripotency markers NANOG (from 1.0 to 0.2), OCT4 (from 1.0 to 0.2), and SOX2 (from 1.0 to 0.3), decreased levels of naïve-state markers KLF4 (from 1.0 to 0.4), FGF4 (from 1.0 to 0.2), NROB1 (from 1.0 to 0.2), and REX1 (from 1.0 to 0.2); and increased mRNA levels of primed-state markers T (from 1.0 to 3.4) and FGF5 (from 1.0 to 6.6) and differentiation genes cTnT (from 1.0 to 3.7, P < 0.05; Fig. 5C), CK8 (from 1.0 to 2.4), BMP4 (from 1.0 to 3.6), DESMIN (from 1.0 to 1.9), PAX6 (from 1.0 to 6.0), and SOX17 (from 1.0 to 4.4, P < 0.05; Supporting Information Fig. S3E). By contrast, compared with PKCi-derived control mES (normalized as 1.0), MBD3 knockdown in mES cultured without PKC inhibitor showed reduced MBD3 mRNA (from 2.1 to 1.1) and protein (from 1.7 to 1.3) levels, albeit its protein still higher than those in PKCi-derived mES (P < 0.05, Fig. 5B). In addition, MBD3 knockdown increased mRNA levels of pluripotent markers NANOG (from 0.2 to 0.5) and SOX2 (from 0.3 to 0.6) and naïve-state markers KLF4 (from 0.4 to 0.5) and NROB1 (from 0.2 to 0.6, P < 0.05); decreased mRNA levels of primed-state markers T (from 3.4 to 1.8) and FGF5 (from 6.6 to 3.4) and differentiation genes cTnT (from 3.7 to 1.5, Fig. 5C), SOX17 (from 4.4 to 2.9), and BMP4 (from 3.6 to 2.2); and recovered mRNA levels of PAX6 to initial PKCi levels (from 6.0 to 2.8; Supporting Information Fig. S3E).
A MBD3 RNA expression increased in mES at passage 5 when the PKC inhibitor was removed for 48 h and was partially reversed by MBD3 knockdown, with the PKCi and PKCi removal plus shNC transfection groups used as controls. B MBD3 and β-actin protein levels were evaluated by Western blot (upper panel). Quantitative density analysis showed that MBD3 protein levels increased after PKC inhibitor was removed, which was partially reversed by MBD3 knockdown (lower panel). C PKC inhibitor removal decreased mRNA levels of pluripotency genes NANOG, OCT4, and SOX2 and naïve-state markers KLF4, FGF4, REX1, and NROB1, whereas MBD3 knockdown partially increased the mRNA levels of pluripotency gene, NANOG, SOX2, KLF4, and NROB1. PKC inhibitor removal increased mRNA levels of primed-state markers T and FGF5 and the mesoderm gene cTnT, whereas MBD3 knockdown partially decreased mRNA levels of T, FGF5, and cTnT. D NANOG, KLF4, FGF5, cTnT, and β-actin levels were detected by Western blot (left panel). Quantitative density analysis showed that NANOG and KLF4 protein levels decreased and FGF5 and cTnT protein levels increased after PKC inhibitor removal, whereas NANOG and KLF4 protein levels partially increased, but FGF5 and cTnT levels partially decreased after MBD3 knockdown (right panel). E After PKC inhibitor removal, mES differentiated and did not show AP staining, whereas MBD3 knockdown partially recovered mES morphology and restored AP staining. Scale bar, 200 μm (upper panel). PKC inhibitor removal decreased the total number of AP-positive colonies, which was partially reversed by MBD3 knockdown (left panel). MBD3 knockdown partially reversed the decrease in the percentage of mixed and undifferentiated colonies and the increase in the percentage of differentiated colonies induced by PKC inhibitor removal (right panel). Data were shown as mean ± SD (n = 3). The letters a, b, and c indicated significant differences among groups (P < 0.05).
Western blot showed that the removal of PKC inhibitor in culture significantly reduced mES protein levels of NANOG (from 1.0 to 0.2) and KLF4 (from 1.0 to 0.4) and increased protein levels of FGF5 (from 1.0 to 6.2) and cTnT (from 1.0 to 3.4) (Fig. 5C). However, MBD3 knockdown in mES cultured without PKC inhibitor partially increased protein levels of NANOG (from 0.2 to 0.4) and KLF4 (from 0.4 to 0.6) and decreased protein levels of FGF5 (from 6.2 to 3.0) and cTnT (from 3.4 to 1.6), but there was not full recovery to initial PKCi levels (P < 0.05; Fig. 5D). Immunostaining showed that after PKC inhibitor removal, mES differentiated into flattened cells that did not express NANOG, whereas knockdown of MBD3 partially prevented mES differentiation and induced weak NANOG expression compared with shNCs cultured without PKC inhibitor (P < 0.05; Supporting Information Fig. S3A–S3D).
PKC inhibitor removal induced mES differentiation, and a few cell colonies existed that were negative for AP staining (Fig. 5E). Quantitative analysis showed that PKC inhibitor removal significantly reduced the number of AP-positive colonies (from 171 to 26), and the percentage of mixed (from 42.5% to 27.4%) and undifferentiated (from 51.3% to 0%) colonies and increased the percentage of differentiated colonies (from 7.2% to 72.6%; Fig. 5E). However, MBD3 knockdown partially recovered mES colony morphology, increased the number of AP-positive colonies (from 26 to 67) and percentage of undifferentiated (from 0% to 7.5%) and mixed (from 27.4% to 52.4%) colonies, and decreased the percentage of differentiated colonies (from 72.6% to 40%, P < 0.05; Fig. 5E).
Discussion
We demonstrated that inhibition of the PKC-signaling pathway by Gӧ6983 reduced the expression of NuRD components at both RNA (MBD3, HDAC1/ HDAC2, MTA1, RbAP46/RbAP48, and p66α) and protein (MBD3 and HDAC2 at passage 3, further MTA1 at passage 10) levels in mES. The PKC-signaling pathway is a complex signal transduction network that participates in other signaling pathways, such as MEK/ERK [20], CREB [21], PKCζ-NF-ĸB [8, 9], PKCµ [19], and GSK3β [22]. Small molecules, such as PD0325901 and CHIR99021 inhibit MEK/ERK and GSK3β pathways, which together with activation of JAK-STAT3 maintain mES naïve pluripotency [5, 7]. In our study, we found comparable ratios of p-ERK/ERK and p-AKT/AKT between 2iL- and PKCi-derived mES, suggesting that inhibition of PKC by the small molecule Gӧ6983 plays a similar role as PD0325901 in suppressing MEK/ERK pathway. On the other hand, we observed an elevated ratio of p-β-catenin/β-catenin in PKCi-derived mES, implying that Gӧ6983 inhibits GSK3-β more effectively than CHIR99021. A previous study [8] reported that GSK3, ERK1/2, AKT, and their respective downstream target genes β-catenin, RSK1, and STAT3 were not phosphorylated in the PKCi system, suggesting that PKCi regulates mES self-renewal independently of traditional signaling pathways. However, we found that the p-ERK/ERK ratio was lower in the PKCi system compared with that after PKC inhibitor removal, indicating that the ERK-signaling pathway is involved in PKCi-derived mES self-renewal. Interestingly, in human ES [23], FGF2 activates PI3K/AKT, MEK/ERK1/2, and PKC isoforms (i.e., PKCδ/ε/ζ), resulting in phosphorylation of GSK-3β, and that activation of AKT signaling promotes self-renewal whereas activation of GSK3β and ERK1/2 induces differentiation. In addition, PKCi system is involved in the downregulation of HDAC5, a key protein in the PKCµ pathway, which maintains mES self-renewal and prevents cell lineage commitment and differentiation, moreover, the inhibition of PKCµ suppresses myoblast differentiation by inhibiting MYOD and myocyte-specific enhancer factor 2C [19]. Key transcription factors are pivotal for maintaining gene expression patterns, chromatin structure, and the epigenetic landscape allowing ES self-renewal [5, 7, 8]. These transcription factors affect the expression of other repressive regulatory factors, including the NuRD complex. Although transcription levels of HDAC1, MTA1, CHD3, RbAP46, and RbAP48 were changed, protein levels were not affected, probably due to the regulation of translation and the lower threshold of RNA required for translation. The mechanism that PKCi down-regulates the expression of MBD3 and other genes is unknown. In the present study, a reduction in NuRD expression was confirmed by decreased expression of NuRD targeted CBR3 and HTRA1 mRNA [1] in PKCi-derived mES. It is assumed that NuRD targeted gene may play a role in regulating the naïve pluripotency.
Previous studies using Gӧ6983, a selective inhibitor of PKC isoforms, show that the PKCζ–NF-ĸB–miRNA-21/miRNA-29 regulatory signaling axis plays a critical role in inducing mES lineage commitment and is capable of maintaining mES and rat ES-specific epigenetic modifications and self-renewal [8, 9]. Our study confirms this finding by showing that knockdown of PKCζ partially rescued the amount of AP-positive mES colonies upon the removal of PKC inhibitor (Fig. S2A, S2B), indicating that inhibition of the NF-ĸB signaling pathway plays an important role in maintaining PKCi-derived mES pluripotency.
The NuRD complex is an abundant and conserved remodeling complex repressor that increases nucleosome density and fine-tunes differential gene expression, even at active transcription sites [1]. MBD3, a key NuRD subunit, is necessary for the development of pluripotent cells, as MBD3-depleted mES are viable but fail to form stable NuRD complexes, which severely affect their cell commitment and differentiation capacities [12]. In the present study, we found that MBD3 plays an important role in mES self-renewal and differentiation. In the PKCi system, knockdown of MBD3 increased the expression of the pluripotency genes NANOG and OCT4, thereby promoting mES self-renewal. Interestingly, knockdown of MBD3 did not affect other pluripotency genes such as KLF4, SOX2, REX1, NROB1, or FGF4. In particular, a previous study reported that KLF4, a naïve-state marker and direct downstream target of LIF/STAT3 [24, 25], mediates the self-renewal-promoting effects of GBX2 in ESs [26]. Therefore, we assume that the LIF/STAT3-signaling pathway does not play a dominant role in the PKCi system. In fact, a previous study found mES devoid of STAT3, suggesting that LIF/STAT3 signaling does not instruct ES self-renewal but may instead act in unrefined culture conditions [7]. By contrast, we observed that MBD3 overexpression and/or PKC inhibitor removal increased MBD3 mRNA and protein levels, downregulated mRNA levels of pluripotency genes such as NANOG and OCT4 and naïve-state genes such as REX1, and upregulated the expression of differentiation genes such as the endoderm marker CK8, mesoderm markers cTnT, BMP4, and DESMIN, and ectoderm markers PAX6 and SOX17. Overexpression of MBD3 also reduced protein levels of NANOG, OCT4, and REX1 but increased protein levels of the primed-state marker FGF5 and differentiation marker cTnT. As a result, the number of ES colonies decreased due to cell differentiation. Furthermore, MBD3 knockdown by RNA interference partially reversed the cell differentiation induced by PKC inhibitor removal. Therefore, our findings support previous findings that the repression of genes related to lineage commitment and cell differentiation is necessary for maintaining mES self-renewal [7, 27]. MBD3 directly regulates pluripotency gene transcription in ESs [16], with one study reporting that MBD3 and Brg1 antagonistically regulate a common set of genes by regulating promoter nucleosome occupancy to maintain pluripotency of embryonic stem cells in mouse [28]. The balance between repressive NuRD and activating chromatin remodeling complex BAF may finely tune gene expression specific to stem cell pluripotency [1]. As a result, mES exhibit a transient cell state poised for cell lineage differentiation during development [5, 29].
In summary, we found that the PKCi culture system reduced the expression of the repressive and epigenetic regulatory NuRD complex and its MBD3, HDAC2, and MTA1 subunits in mES. This reduction in NuRD promoted the expression of pluripotency genes that maintain the naïve state of mES. Functional knockdown and overexpression experiments revealed that MBD3, a key NuRD subunit, plays an important role in regulating the expression of genes responsible for mES pluripotency. This demonstration of the transcriptional/translational regulatory effects of the NuRD complex on a wide spectrum of developmental genes increases our understanding of ES pluripotency, lineage commitment, and differentiation.
Materials and methods
Chemicals and reagents
Unless otherwise stated, all chemicals and reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Animal maintenance, hormone-induced superovulation, and blastocyst collection
Animal experimental protocols were approved by the Animal Care and Use Committee of Nanjing Normal University (NSD-2013-30) and were performed according to guidelines from the US National Institutes of Health. At 6–8 weeks of age, female C57BL/6J mice were intraperitoneally injected with 7.5 IU pregnant mare serum gonadotropin (Ningbo Second Hormone Factory, China). Forty-eight hours later, mice were intraperitoneally injected with 7.5 IU human chorionic gonadotropin (Ningbo Second Hormone Factory) and mated with males (1:1). Blastocysts were flushed from the uterus with M2 medium on day 3.5.
De novo derivation of mES
Blastocysts were seeded on Mitomycin C-treated mouse embryonic fibroblasts on 0.1% gelatin-coated plates (ES-006-B, Millipore, USA) with PKC inhibitor (5 μM Gӧ6983, 133053-19-7, Selleck, USA) in basic culture medium supplemented with Dulbecco’s modified Eagle’s medium (DMEM; 10829018, Gibco, USA) containing 15% knockout serum replacement (10828028, Gibco), 1% penicillin/streptomycin (SV30010, HyClone, USA), 2 mM glutamine (35050061, Gibco), 1 mM sodium pyruvate (11360088, Gibco), and 0.1 mM 2-mercaptoethanol (ES-007-E, Millipore). After culture for 7 days, outgrowths were collected and digested into single-cell suspensions with accutase (A1110501, Gibco) and re-seeded in new plates coated with feeder cells. mES passaging was performed by incubating colonies with accutase, followed by plating at a density of 1 × 103 cells/cm2 into a new 24-well plate coated with new feeder cells at 3- to 4-day intervals. Collected mES were frozen in a cryopreservation medium with 90% fetal bovine serum (FBS; SH30070.03, HyClone) and 10% dimethyl sulfoxide (D5879) and stored in liquid nitrogen.
MBD3 overexpression in mES
FUW-MBD3 (#52356) and control FUW-M2rtTA (#20342) were purchased from Addgene. Lentiviral infection and overexpression were performed as previously described [30]. Briefly, 293T cells were cultured for 2 days in DMEM (C11995500BT, Gibco) supplemented with 10% FBS (v/v) at 37 °C in 5% CO2. At 70–80% confluency, cells were transfected with FUW-MBD3 or control FUW-M2rtTA along with the viral packaging plasmids psPAX and pMD2.G (5:3:2) with Lipofectamine 2000 reagent (1947415, Invitrogen, USA) at a 1:2 ratio of DNA (g) to Lipofectamine 2000 (μL). After 6 h, the medium was changed. After transfection for 48 h, viral supernatants were collected and filtered (0.45-µm pore size; Millipore) to infect mES. For lentiviral transfection, mES at 70–80% confluency were infected with filtered viral supernatants (FUW-MBD3 or FUW-M2rtTA) supplemented with an equal volume of fresh PKCi medium. Twelve hours later, mES were repeatedly infected with viral supernatants up to four times within 48 h as needed.
RNA interference in mES
Lentiviral supernatants containing short hairpin RNA (shRNA) targeting mouse MBD3 mRNA (shMBD3), PKCζ KD1, or PKCζ KD2 or an shRNA negative control (shNC) were purchased from GenePharma (Shanghai, China). The shPKCζ KD1 target sequence was GGGACGAAGTGCTCATCATTC, shPKCζKD2 was GGATCGACCAGTCCGAATTTG, shMBD3 was GCCTCCTTATCATAGGACAAG, and shNC was TTCTCCGAACGTGTCACGT. RNA interference was performed according to the manufacturer’s instructions. Briefly, mES were cultured for 1–2 days with PKCi medium, after which lentiviral diluent was added to the culture medium and incubated overnight at 37 °C in 5% CO2. After 24 h of incubation, the medium was changed to fresh PKCi medium, and incubation continued for another 24 h at 37 °C in 5% CO2. Infected mES were cultured in PKCi-free ES medium for 48 h and collected for further analysis and Western blot assay. shNC infection was used as a knock-down control (−PKCi + shNC).
qPCR
Total RNA was extracted from mES with Trizol reagent (T9424). Reverse transcription reactions were performed with 1 μg RNA using HiScript II Reverse Transcriptase (R223-01, Vazyme, China). Complementary DNA was used as a template, and 2xSYBR Green Fast qPCR Mix with High Rox (RM21206, ABclonal, China) was used for the qPCR reaction. The qPCR primers are shown in Table 1. Individual gene expression was normalized to GAPDH expression, and values from the PKCi group were defined as 1.0 for all gene expression levels.
Western blot
mES total protein was collected with lysis buffer and quantified using a BCA Kit (GK5012, Beyotime Biotechnology, China). Western blot analysis was performed as previously described [31]. Briefly, 6 μg total protein was loaded into each well of a 12% gel for SDS–PAGE. After electrophoresis, separated proteins were transferred to a polyvinylidene fluoride membrane (03010040001, Roche, Basel, Switzerland) by electrotransfer. Membranes were blocked with 5% non-fat powdered milk (A600669, Sangon Biotech, China) for 1 h, after which membranes were washed with tris-buffered saline containingTween 20 (TBST; 9005-64-5, Sangon Biotech) and incubated with anti-p-β-catenin (1:1000, 9561T, Cell Signaling Technology, CST, USA), anti-β-catenin (1:1000, A19657, ABclonal, China), anti-p-ERK (1:1000, 3510, CST), anti-ERK (1:1000, 4695, CST), anti-p-AKT (1:1000, AP1214, ABclonal), anti-AKT (1:1000, A17909, ABclonal), anti-HDAC5 (A0632, ABclonal), anti-HDAC1 (1:1000, A0238, ABclonal), anti-HDAC2 (1:1000, A2084, ABclonal), anti-MTA1 (1:1000, A16085, ABclonal), anti-MBD3 (1:1000, A2251, Lot 0046180101, ABclonal), anti-CHD3 (1:1000, A2221, ABclonal), anti-RbAP46 (1:1000, A6967, ABclonal), anti-RbAP48 (1:1000, A13934, ABclonal), anti-NANOG (1:1000, Lannuo Biotechnology, China), anti-OCT4 (1:1000, Lannuo Biotechnology), anti-REX1 (1:1000, Lannuo Biotechnology), anti-FGF5 (1:1000, ab88118, Abcam), or anti-cTnT (1:1000, A4914, ABclonal) antibodies overnight at 4 °C. The next day, membranes were washed with TBST and incubated with horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (1:5000, BS13278, Bioworld) for 1 h. After washing, membranes were processed using an enhanced chemiluminescence reagent (E411-04, Vazyme), and protein bands were visualized using a LAS-4000 imager (Tanon, Shanghai, China). β-actin (1:1000, AC026, ABclonal) was used as an internal control. Target protein expression levels were normalized to those of β-actin. Values from the PKCi group were defined as 1.0 and were used for comparisons with other treatments.
Immunocytochemical staining for pluripotency markers
Immunostaining experiments were performed with the pluripotency marker NANOG (1:200, Lannuo Biotechnologies). Briefly, mES were washed with Dulbecco’s phosphate-buffered saline (DPBS) three times, fixed with 4% paraformaldehyde for 10 min at room temperature, washed with DPBS three times, incubated with 0.2% Triton-X 100 (Solarbio, China) for 5 min, washed with DPBS three times, incubated with 2% FBS (HyClone) for 30 min at room temperature to block nonspecific binding, and incubated with primary antibodies at 4 °C overnight. The next day, cells were washed with DPBS three times, incubated with secondary antibody conjugated with FITC (goat anti-rabbit IgG (H + L), 1:300, AS011, ABclonal) for 2 h at room temperature, washed with DPBS, and stained with 100 ng/ml DAPI (SunShine, China) for 10 min at room temperature in the dark. Finally, after washing with DPBS, cells were observed under an inverted fluorescence microscope.
Alkaline phosphatase staining
mES were washed with DPBS three times, fixed with 4% paraformaldehyde for 10 min at room temperature, washed with DPBS three times, incubated with nitro blue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate substrates (REF 11745832910, Roche, Switzerland) for 20–30 min at room temperature, washed with DPBS, and observed under an inverted microscope.
Statistical analysis
All experiments were repeated at least three times. mES derived from either PKCi or LIF-2i were from different blastocysts, but mES after derivation in two conditions were the same strains to exclude the possibility of variations. Data were analyzed using SPSS version 18.0 (SPSS, Inc., Chicago, IL, USA) and were shown as mean ± standard deviation (SD). Statistical comparisons were performed using analysis of variance (ANOVA). The letters a, b, and c indicated significant differences among groups (P < 0.05).
Data availability
All data related to this paper may be requested from the corresponding author.
References
Bornelov S, Reynolds N, Xenophontos M, Gharbi S, Johnstone E, Floyd R, et al. The nucleosome remodeling and deacetylation complex modulates chromatin structure at sites of active transcription to fine-tune gene expression. Mol Cell. 2018;71:56–72. e4
Xue Y, Wong J, Moreno GT, Young MK, Côté J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–61.
Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–89.
Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature 1981;292:154–6.
Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–92.
Silva J, Smith A. Capturing pluripotency. Cell 2008;132:532–6.
Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, et al. The ground state of embryonic stem cell self-renewal. Nature 2008;453:519–23.
Dutta D, Ray S, Home P, Larson M, Wolfe MW, Paul S. Self-renewal versus lineage commitment of embryonic stem cells: protein kinase C signaling shifts the balance. Stem Cells. 2011;29:618–28.
Rajendran G, Dutta D, Hong J, Paul A, Saha B, Mahato B, et al. Inhibition of protein kinase C signaling maintains rat embryonic stem cell pluripotency. J Biol Chem. 2013;288:24351–62.
Nishizuka Y. Protein kinase C and lipid signaling for sustained cellular responses. FASEB J. 1995;9:484–96.
Hu G, Wade PA. NuRD and pluripotency: a complex balancing act. Cell Stem Cell. 2012;10:497–503.
Kaji K, Nichols J, Hendrich B. Mbd3, a component of the NuRD co-repressor complex, is required for development of pluripotent cells. Development 2007;134:1123–32.
Torchy MP, Hamiche A, Klaholz BP. Structure and function insights into the NuRD chromatin remodeling complex. Cell Mol Life Sci. 2015;72:2491–507.
Zhang Y, Huck-Hui N, Hediye E-B, Paul T, Adrian B, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–35.
Kaji K, Caballero IM, MacLeod R, Nichols J, Wilson VA, Hendrich B. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat Cell Biol. 2006;8:285–92.
Reynolds N, Latos P, Hynes-Allen A, Loos R, Leaford D, O’Shaughnessy A, et al. NuRD suppresses pluripotency gene expression to promote transcriptional heterogeneity and lineage commitment. Cell Stem Cell. 2012;10:583–94.
Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell 2007;128:747–62.
Dai YJ, Li JS, Li MY, Liu ZH, Liu J, An LY, et al. Methyl-CpG-binding domain 3 (Mbd3) is an important regulator for apoptosis in mouse embryonic stem cells. Am J Transl Res. 2020;12:8147–61.
Jama A, Huang D, Alshudukhi AA, Chrast R, Ren H. Lipin1 is required for skeletal muscle development by regulating MEF2c and MyoD expression. J Physiol. 2019;597:889–901.
Kunath T, Saba-El-Leil MK, Almousailleakh M, Wray J, Meloche S, Smith A. FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development 2007;134:2895–902.
Wu D, Wu F, Lin R, Meng Y, Wei W, Sun Q, et al. Impairment of learning and memory induced by perinatal exposure to BPA is associated with ERalpha-mediated alterations of synaptic plasticity and PKC/ERK/CREB signaling pathway in offspring rats. Brain Res Bull. 2020;161:43–54.
Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63.
Kinehara M, Kawamura S, Tateyama D, Suga M, Matsumura H, Mimura S, et al. Protein kinase C regulates human pluripotent stem cell self-renewal. PLoS ONE. 2013;8:e54122.
Hall J, Guo G, Wray J, Eyres I, Nichols J, Grotewold L, et al. Oct4 and LIF/Stat3 additively induce Kruppel factors to sustain embryonic stem cell self-renewal. Cell Stem Cell. 2009;5:597–609.
Niwa H, Ogawa K, Shimosato D, Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature 2009;460:118–22.
Wang M, Tang L, Liu D, Ying QL, Ye S. The transcription factor Gbx2 induces expression of Kruppel-like factor 4 to maintain and induce naive pluripotency of embryonic stem cells. J Biol Chem. 2017;292:17121–8.
Xu X, Guo M, Zhang N, Ye S. Telomeric noncoding RNA promotes mouse embryonic stem cell self-renewal through inhibition of TCF3 activity. Am J Physiol Cell Physiol. 2018;314:C712–C20.
Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS, et al. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell 2011;147:1498–510.
Takahashi S, Kobayashi S, Hiratani I. Epigenetic differences between naive and primed pluripotent stem cells. Cell Mol Life Sci. 2018;75:1191–203.
Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, Cook EG, et al. Parkinson’s disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell 2009;136:964–77.
Hayashi Y, Furue MK, Okamoto T, Ohnuma K, Myoishi Y, Fukuhara Y, et al. Integrins regulate mouse embryonic stem cell self-renewal. Stem Cells. 2007;25:3005–15.
Acknowledgements
This study was supported in part by the Natural Science Foundation of China (NSFC) (Grant Nos. 31872353, 32072732, 31340041, and 31471388) and Priority Academic Program Development of Jiangsu Higher Education Institutions to FD and the National Key R&D Program of China (2018YFC1004500), The Stable Support Plan Program of Shenzhen Natural Science Fund (20200925153547003), Shenzhen Innovation Committee of Science and Technology (ZDSYS20200811144002008), and Southern University of Science and Technology (No.G02226301 and Y01501821) to CH.
Author information
Authors and Affiliations
Contributions
FD, CH, LA, and YD designed the experiments; YD performed the experiments; JS, NH, and CH analyzed the experimental data; FD and YD wrote the manuscript; CH and FD discussed the manuscript arrangement.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Dai, Y., Sun, J., He, N. et al. Nucleosome remodeling and deacetylation complex and MBD3 influence mouse embryonic stem cell naïve pluripotency under inhibition of protein kinase C. Cell Death Discov. 8, 344 (2022). https://doi.org/10.1038/s41420-022-01131-0
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41420-022-01131-0
This article is cited by
-
Characterization of a Distinct State in the Continuum of Pluripotency Facilitated by Inhibition of PKCζ in Mouse Embryonic Stem Cells
Stem Cell Reviews and Reports (2023)