Abstract
Dysregulated long noncoding RNAs (lncRNAs) are involved in the pathogenesis and development of human diseases, such as epithelial ovarian cancer (EOC). In this study, we identified EOC-related lncRNAs and performed lncRNA and mRNA microarray analyses using IOSE80, a normal ovary cell line, and two ovarian carcinoma cell lines (SKOV3 and SKOV3/DDP) to investigate the potential roles of lncRNAs in EOC. lncRNA-HEIH expression in EOC tissues and cell lines was measured by quantitative real-time polymerase chain reaction (qPCR). In addition, we generated a lncRNA–mRNA co-expression network in order to identify lncRNA-expression trends and potential lncRNA target genes. Cell viability, migration, and invasion were determined by Cell Counting Kit-8, transwell assay, and wound-healing assay, respectively, and apoptosis was analyzed by flow cytometry. We identified 3527 differentially expressed lncRNAs upon comparison of the lncRNA profiles from IOSE80 with those of SKOV3 cell lines, with 11 differentially expressed lncRNAs confirmed by qPCR. Both pathway and gene ontology analyses demonstrated the involvement of lncRNAs, especially HEIH and LINC-PINT, in multiple biological processes. Furthermore, in vitro knockdown experiments confirmed that suppression of HEIH expression inhibited EOC cell proliferation. Our findings provide a foundation for further research into the role of these lncRNAs in EOC carcinogenesis and progression.
Similar content being viewed by others
Introduction
Ovarian cancer (OC) is the third most common cancer in women and the most fatal gynecological cancer in the world. According to a recent study, 22,280 new cases of OC and 15,500 OC-related deaths have been reported in the United States1. Epithelial ovarian cancer (EOC) constitutes 90% of all OC cases and is typically diagnosed at an advanced stage due to rapid progression of the disease and nonspecific clinical symptoms. Despite recent progress in surgery and chemotherapy, the overall 5-year survival rate of EOC patients is only 40%2. Due to the poor understanding of EOC etiology and pathogenesis, the molecular mechanisms associated with EOC progression have not been clearly elucidated, which is critical for the development of new therapeutic and diagnostic approaches.
Long noncoding RNAs (lncRNAs) are nonprotein-coding RNAs of >200 nucleotides and involved in the regulation of gene expression3. lncRNA-expression levels exhibit high degrees of tissue and disease specificity, which are closely associated with their biological functions4. Recent studies show that dysregulated lncRNAs play vital roles in tumorigenesis5 via multiple cancer-related biological processes, including apoptosis, cell cycle regulation, metastasis, and DNA-damage response6,7,8. Several lncRNAs, such as HOTAIR9, FAL110, and HOST211, are involved in EOC progression, with ZFAS1 upregulated by cisplatin in multiple EOC cell lines12. In addition, a peptide nucleic acid-based therapeutic approach decreased EOC invasiveness and increased chemotherapeutic sensitivity by inhibiting HOTAIR–EZH2 activity13. Moreover, the lncRNA FAL1 regulates cancer-cell cycle progression and cell senescence, as well as xenograft tumor growth in vivo14, and another study reported that inhibiting the expression of lncRNA HOST2 significantly reduced the migration, invasion, and proliferation of OVCAR-3 cells15. However, the regulatory roles of lncRNAs in EOC have not been studied sufficiently.
In this study, we performed genome-wide lncRNA and mRNA microarray analyses on IOSE80 cells, a normal ovary cell line, and two OC cell lines (SKOV3 and SKOV3/DDP) in order to identify EOC-related lncRNAs. The SKOV3 cell line is a human OC adenocarcinoma cell line, and SKOV3/DDP is a cisplatin-resistant variant of SKOV316. Our findings revealed functional lncRNA profiles in EOC and cis-resistant EOC cells, thereby expanding the current understanding of EOC pathogenesis and providing novel insights supporting the development of new therapeutic targets for EOC.
Results
Determination of lncRNA-expression profiles in different cell lines
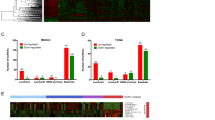
We profiled IOSE80, SKOV3, and SKOV3/DDP cell lines in order to determine transcriptional alterations. A total of 40,173 lncRNAs were analyzed using the Arraystar human lncRNA microarray version 4.0, and differences in lncRNA-expression levels according to fold changes >2.00 and a P < 0.05 were considered significant. Hierarchical clustering showed distinguishable lncRNA-expression profiles among the different cell lines, with the results indicating 3527 differentially expressed lncRNAs between SKOV3 and IOSE80 cells, of which 1945 were upregulated and 1582 were downregulated. A total of 9706 differentially expressed lncRNAs were identified in the SKOV3/DDP cell line as compared with the IOSE80 cell line and included 6205 upregulated and 3501 downregulated lncRNAs. Moreover, the expression profiles of 9314 lncRNAs differed between SKOV3/DDP and SKOV3 cells, including 5748 upregulated and 3566 downregulated lncRNAs. Figure 1 shows volcano plots and hierarchical clustering of the differentially expressed lncRNAs.
a Differentially expressed lncRNAs were analyzed using hierarchical clustering. ‘Red’ indicates high relative expression, and ‘green’ indicates low relative expression. Volcano plot comparisons of gene expression between b SKOV3 and IOSE80 cells, c SKOV3/DDP and IOSE80 cells, and d SKOV3/DDP and SKOV3 cells. Vertical lines correspond to twofold upregulation and downregulation, respectively, and the horizontal line represents a P < 0.05. The red and green points in the plot represent upregulated and downregulated genes that are statistically significant
mRNA profiles of the different cell lines
We performed microarray profiling of mRNAs, identifying a total of 5497 differentially expressed mRNAs between SKOV3 and IOSE80 cells, including 2320 upregulated and 3177 downregulated mRNAs. In addition, 12,015 mRNAs were differentially expressed between SKOV3/DDP and IOSE80 cells, including 5336 upregulated and 6679 downregulated mRNAs. Moreover, we identified 11,587 differentially expressed mRNAs between SKOV3/DDP and SKOV3 cells, including 5328 upregulated and 6259 downregulated mRNAs. Figure 2 shows volcano plots and hierarchical clustering results for the differentially expressed mRNAs.
a Differentially expressed mRNAs were analyzed using hierarchical clustering. Volcano plot comparisons of gene expression between b SKOV3 and IOSE80 cells, c SKOV3/DDP and IOSE80 cells, and d SKOV3/DDP and SKOV3 cells. Volcano plots were used to visualize differential expression between two different conditions. Vertical lines correspond to twofold upregulation and downregulation, respectively, and the horizontal line represents a P < 0.05. The red and green points in the plot represent upregulated and downregulated genes that are statistically significant
Biological analysis
In our survey of gene ontology (GO) biological processes, the functions of the upregulated mRNAs (SKOV3 versus IOSE80, SKOV3/DDP versus IOSE80, and SKOV3/DDP versus SKOV3) were primarily involved in anatomical structure development, single-organism processes, and regulation of cellular biosynthetic processes (Fig. 3a–c). In addition, the functions of the downregulated mRNAs primarily involved cellular metabolic processes, cellular macromolecule metabolic processes, and cellular response to interferon (IFN)-γ (Fig. 3d–f).
a–c Top ten GO terms of biological processes for mRNAs downregulated between SKOV3 and IOSE80 cells, SKOV3/DDP and IOSE80 cells, and SKOV3/DDP and SKOV3 cells. d–f Top ten GO terms of biological processes for mRNAs upregulated between SKOV3 and IOSE80 cells, SKOV3/DDP and IOSE80 cells, and SKOV3/DDP and SKOV3 cells
The main Kyoto encyclopedia of genes and genomes (KEGG) pathways for the upregulated transcripts (SKOV3/DDP versus IOSE80, SKOV3/DDP versus SKOV3, and SKOV3 versus IOSE80) involved endocytosis, the mitogen-activated protein kinase (MAPK) signaling pathway, and the p53 signaling pathway (Fig. 4a–c). By contrast, the main KEGG pathways for the downregulated transcripts included ribosome, protein processing in the endoplasmic reticulum, antigen processing and presentation, and herpes simplex infection (Fig. 4d–f).
a–c Top ten KEGG pathways corresponding to the mRNAs downregulated between SKOV3 and IOSE80 cells, SKOV3/DDP and IOSE80 cells, and SKOV3/DDP and SKOV3 cells. d–f Top ten KEGG pathways corresponding to the mRNAs upregulated between SKOV3 and IOSE80 cells, SKOV3/DDP and IOSE80 cells, and SKOV3/DDP and SKOV3 cells
Candidate lncRNA-expression levels in vivo
Based on the microarray results of the 3527 lncRNAs, we identified four with the most significant differential expression between OC and normal ovary cells (CDKN2A-AS1, LINC00184, LINC-PINT, and LOC100133669). We performed real-time quantitative polymerase chain reaction (qPCR) analysis of the levels of these lncRNAs in the cell lines described, as well as a new ovary cancer cell line (HO-8910), in order to verify their expression. The results demonstrated that CDKN2A-AS1 and LINC00184 were significantly upregulated, whereas LINC-PINT and LOC100133669 were markedly decreased in HO-8910, SKOV3, and SKOV3/DDP cells as compared with levels in IOSE80 cells (Fig. 5a). To confirm these findings, ovary biopsy samples extracted during gynecological surgery from both OC patients and healthy individuals were used to evaluate differences in levels of the most dysregulated candidate lncRNAs, including CDKN2A-AS1, C7orf55, LINC00184, HEIH, ZEB2, SNHG3, LINC00630, LOC100133669, APTR, LINC-PINT, and SEMA5A. qPCR results indicated marked upregulation of CDKN2A-AS1, C7orf55, LINC00184, HEIH, and ZEB2 expression, whereas that of SNHG3, LINC00630, LOC100133669, APTR, LINC-PINT, and SEMA5A was downregulated in OC tissues as compared with levels in normal ovary tissues (Fig. 5b).
Construction of a lncRNA–mRNA co-expression network
A lncRNA–mRNA co-expression network was constructed in order to identify mRNAs associated with specific lncRNAs. The two lncRNAs exhibiting the most significant differential expression (HEIH and LINC-PINT) were selected for building this co-expression network in order to evaluate potential associations with mRNAs. We selected 74 and 5 target mRNAs for HEIH and LINC-PINT, respectively, among which were those involved in important roles in cancer progression. For example, S-phase kinase-associated protein 2 participates in the regulation of immune reactions, cell proliferation, and recruitment, and levels of schlafen family member 11 are directly associated with chemotherapeutic and/or polyADP ribose polymerase-inhibitor sensitivity in a number of cancer types. In addition, upregulation of MAN1A1 activates the unfolded protein response and might initiate metastasis. The cancer-specific roles of these mRNAs suggest the potential importance of HEIH and LINC-PINT in EOC (Fig. 6a, b).
Suppression of lncRNA HEIH inhibits OC cell progression
To further investigate the function of HEIH in OC development, we transfected SKOV3 and HO-8910 cells with small-interfering (si)RNA targeting HEIH (si-HEIH), followed by assessment of cell proliferation, migration, and invasion. Both OC cell lines exhibited lower invasion rates relative to those of control cells, suggesting that HEIH silencing significantly reduced the invasive abilities of SKOV3 and HO-8910 cells (Fig. 7a, c, h, j). To evaluate the role for HEIH in cell migration, we performed a wound-healing assay, finding that HEIH-silenced cells exhibited slower wound-healing ability relative to controls (Fig. 7b, d, i, k). In addition, flow cytometry results showed a decreased number of cells in the S phase of the cell cycle (Fig. 7e, f, l, m). Moreover, Cell Counting Kit-8 (CCK-8) assays confirmed that HEIH silencing inhibited OC cell viability (Fig. 7g, n). These results suggested that HEIH siRNA might represent an efficacious method for suppressing tumor growth by inhibiting OC cell progression.
SKOV3 and HO-8910 cells were transfected with si-HEIH for 24 h. a, h SKOV3 and HO-8910 invasion abilities according to the transwell migration assay. b, i Representative photomicrographs of wound healing in the presence of SKOV3 cells at 0 h and 24 h. c, j Number of SKOV3 and HO-8910 cells that invaded the substratum of the membrane per field of view. d, k Photomicrographs of wound healing in the presence of HO8910 at 0 h and 24 h. Arrows highlight the linear scratch/wound for each group of cells. The mean percentage of wound closure per group (n = 3). e, f, l, m Flow cytometric results showing cell cycle arrest in the si-HEIH group. g, n Proliferative ability according to the CCK-8 assay. Data represent the mean ± standard deviation. *P < 0.05; **P < 0.01; ***P < 0.001
Discussion
Numerous studies report that lncRNA dysregulation is associated with human cancers, including EOC17. However, studies of the expression profiles of lncRNAs in EOC or cis-resistant EOC, as well as those predicting correlations between lncRNA and malignant progression, are limited. In this study, we used microarrays to detect genome-wide lncRNA-expression patterns in SKOV3 cell lines and compared those with patterns in IOSE80 and SKOV3/DDP cell lines in order to elucidate their potential roles in EOC oncogenesis and progression. In addition, we evaluated correlated signaling pathways associated with several candidate lncRNAs identified from this analysis.
lncRNAs play important roles in the regulation of gene expression and tumor progression18,19, and their expression is closely associated with biological functions and tumor status20. Some lncRNAs exhibit tissue-specific, disease-specific, and developmental-stage-specific expression20,21,22. Previous studies identified HOTAIR23, ANRIL24, and LSINCT525 as important regulators of OC. In the present study, we observed significant differential expression of some lncRNAs and mRNAs in SKOV3 and SKOV3/DDP cells as compared with IOSE80 cells, and that a higher number of genes exhibited differential expression in SKOV3/DDP cells relative to SKOV3 cells. Our results highlighted a unique lncRNA-expression profile in SKOV3 cells, with some of these lncRNAs possibly associated with chemotherapy resistance.
We found that most of the enriched GO terms and KEGG pathways associated with downregulated mRNAs were shared between SKOV3 and SKOV3/DDP cells. GO analysis revealed that differentially expressed lncRNAs were highly enriched in cellular response to IFN-γ, cellular metabolic processes, single-organism processes, and regulation of cellular biosynthetic processes. Moreover, the top predicted KEGG pathways were the p53 signaling pathway, cancer-related pathways, protein-processing pathways in the endoplasmic reticulum, and the MAPK signaling pathway, all of which are associated with EOC progression. Previous studies showed that lncRNAs play important roles in cancer by regulating associated signaling pathways, with PINCR26, MIR31HG27, and ROR28 affecting cancer-cell proliferation via the p53 signaling pathway, and NNT-AS129 and BANCR30 influencing cancer-cell proliferation and invasion by regulating the MAPK signaling pathway. Our results indicated that dysregulated lncRNAs might regulate cancer-associated pathways and affect the pathogenic process of EOC. Moreover, the significantly enriched GO terms and KEGG pathways for upregulated RNAs between the SKOV3 and SKOV3/DDP cell lines differed considerably from those of the downregulated RNAs. Our findings suggested that lncRNAs potentially regulate chemotherapy resistance via the enhancement of particular signaling pathways.
Our findings indicated that the functions of most of the identified lncRNAs were ambiguous. We constructed a lncRNA–mRNA co-expression network in order to identify key lncRNAs associated with EOC. lncRNAs HEIH and LINC-PINT showed the highest number of neighbors in the network, suggesting potentially key roles in regulating gene expression and protein translation possibly involved in EOC progression. Previous studies reported that a conserved functional codependence between LINC-PINT and polycomb repressive complex 2 counteracts gene activation by the early growth response-1 protein31, and that HEIH promotes melanoma-cell proliferation, migration, and invasion by inhibiting miR-200b/a/429 levels32. In addition, we found that part of the dysregulated lncRNAs in the SKOV3/DDP cell line showed a higher degree of significant differential expression relative to those in the SKOV3 cell line. Moreover, CCK-8 and flow cytometric analyses confirmed that HEIH silencing significantly reduced the proliferative capacity of OC cells, and wound-healing and transwell migration assays indicated key roles for HEIH in inhibiting OC cell migration and invasion.
In summary, our findings demonstrated that 8388 lncRNAs were differentially expressed in SKOV3 cell lines as compared with the IOSE80 cell line, and that multiple lncRNAs were identified as potentially involved in EOC-related signaling pathways. Our data provides a foundation for further investigations into the roles of HEIH in EOC oncogenesis and progression and will promote identification of novel therapeutic targets and diagnostic biomarkers in the future.
Materials and methods
Cell culture
IOSE80, SKOV3, and SKOV3/DDP cell lines were purchased from the American Type Culture Collection (Manassas, VA, USA). The HO-8910 human OC cell line was obtained from the Department of Obstetrics and Gynecology Laboratory of the Second Affiliated Hospital of Harbin (Heilongjiang, China), and Roswell Park Memorial Institute (RPMI)-1640 medium was purchased from Hyclone (South Logan, UT, USA). IOSE80, SKOV3, and SKOV3/DDP cells were cultured in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum (FBS). HO-8910 cells were cultured in RPMI-1640 medium supplemented with 10% FBS. All cell lines were cultured in a humidified incubator in an atmosphere of 5% CO2 at 37 °C.
lncRNA and mRNA microarray
The Arraystar human lncRNA microarray (v.4.0; Arraystar, Rockville, MD, USA) was used for the global expression profiling of human lncRNAs and protein-coding mRNA transcripts in the EOC cell lines. Total RNA was amplified and labeled using a low-input QuickAmp labeling kit (One-Color; Agilent Technologies, Santa Clara, CA, USA). Labeled complementary (c)RNA was purified using an RNeasy mini kit (Qiagen, Hilden, Germany). The concentration and specific activity of the labeled cRNAs [pmol cyanine 3 (Cy3)/μg cRNA] were measured using a NanoDrop ND-1000 (Thermo Fisher Scientific, Waltham, MA, USA). Each slide was hybridized with 1.65 μg Cy3-labeled cRNA for 17 h at 65 °C in an Agilent hybridization oven using the Agilent gene-expression hybridization kit (No. 5188–5242; Agilent Technologies). The slides were washed in a slide-staining dish (Shandon; No. 121; Thermo Fisher Scientific) using a gene-expression wash buffer kit (Agilent Technologies) and scanned using an Agilent microarray scanner (G2565BA; Agilent Technologies). Feature-extraction software (v.11.0.1.1; Agilent Technologies) was used to extract data and analyze the acquired array images. Microarray data were saved in the Gene Expression Omnibus database (accession ID: GSE104776).
RNA isolation and real-time qPCR
Total RNA was extracted from cells using TRIzol reagent (Thermo Fisher Scientific), with the amount of TRIzol reagent used based on the area of the culture dish (1 mL/10 cm2). Microarray data were validated by qPCR using SYBR Green master mix (Applied Biosystems, Foster City, CA, USA). β-actin was used as an internal control, and the 2−ΔΔCT method was used for transcript quantification. Primers were designed using Primer 5 software (Premier Biosoft, Palo Alto, CA, USA), with the primer sequences listed in Table 1. All experiments were independently performed in triplicate.
Pathway analysis
Using the latest KEGG database (http://www.genome.jp/kegg), we performed pathway analyses for the differentially expressed mRNAs in order to determine significantly enriched biological pathways according to P < 0.05.
GO analysis
We performed GO analysis in order to associate differentially expressed mRNAs with categories from the GO website (http://www.geneontology.org) according to P < 0.05, which denoted a significant GO term.
lncRNA–mRNA co-expression network
We constructed a lncRNA–mRNA co-expression network in order to investigate the potential functions of the differentially expressed lncRNAs via interactions between mRNAs and lncRNAs. Networks were constructed using a previously described algorithm33. In the networks, red nodes represented mRNAs, and green nodes represented lncRNAs, with the lines between the red and green nodes representing interactions between mRNAs and lncRNAs. A solid line indicated a positive correlation, whereas a dashed line indicated a negative correlation.
Transwell migration assay
A transwell migration assay was performed using Transwell inserts (BD Biosciences, San Jose, CA, USA) with an 8-μm pore-size filter. A total of 5 × 104 cells in serum-free medium were seeded into the upper chamber of the insert pre-coated with Matrigel, and 500 μL of conditioned medium was added to the lower chamber. After a 24-h incubation, the cells were fixed with paraformaldehyde for 15 min and stained with 0.1% crystal violet, after which the cells on the top surface of the membrane were carefully removed, and the cells on the lower surface were examined by microscopy. Five random fields were photographed for counting purposes, and the average number of migrated cells was used as an indicator of migration capacity. All experiments were independently performed in triplicate.
Cell-proliferation assay
Cell-proliferative capacity was measured using CCK-8 (Dojindo, Shanghai, China). Briefly, 2.5 × 103 cells were seeded into wells after transfection and cultured for 24 h, after which 10 μL of CCK-8 reagent was added to each well, followed by incubation for 1 h at 37 °C. Optical density was measured using a microplate reader (Bio-Rad, Hercules, CA, USA) at a wavelength of 450 nm, and proliferation rates were calculated.
Cell scratch test
SKOV3 and HO-8910 cells were seeded at 1 × 105 cells/well on 1% gelatin-coated six-well plates (Corning, Apeldoorn, Netherlands). Confluent cells were serum-deprived for 16 h, and linear wounds were created in the monolayers by scratching with a sterile pipette tip. The monolayers were then washed with phosphate-buffered saline to remove floating cells, followed by addition of the conditioned medium. After a 24-h incubation, cell migration into the wound was assessed by microscopy. All experiments were independently performed in triplicate.
Flow cytometry
We performed flow cytometric analysis in order to determine whether silencing HEIH inhibited the growth phase of SKOV3 and HO-8910 cells. SKOV3 and HO-8910 cells transfected with either negative control siRNA (si-NC) or si-HEIH were harvested at 48 h and resuspended at 1 × 106 cells/mL, followed by the addition of Annexin V-FITC and propidium iodide (PI; BD Pharmingen, Franklin Lakes, NJ, USA) according to manufacturer instructions. Cells were fixed with 70% ice-cold ethanol and labeled with PI for growth-phase analysis. All experiments were conducted using a flow cytometer (FACScan; BD Biosciences).
Statistical analysis
Data represent the mean ± standard error of the mean. Differences among groups were analyzed using one-away analysis of variance accompanied by a Newman–Keuls multiple-comparison test using GraphPad Prism software (v.5.0; GraphPad, San Diego, CA, USA). A two-tailed Student’s t test was used for comparison between two groups. A P < 0.05 was considered statistically significant.
References
Oza, A. M. et al. Standard chemotherapy with or without bevacizumab for women with newly diagnosed ovarian cancer (ICON7): overall survival results of a phase 3 randomised trial. Lancet Oncol. 16, 928–936 (2015).
Gloss, B. S. et al. Integrative genome-wide expression and promoter DNA methylation profiling identifies a potential novel panel of ovarian cancer epigenetic biomarkers. Cancer Lett. 318, 76–85 (2012).
Dykes, I. M. & Emanueli, C. Transcriptional and post-transcriptional gene regulation by long noncoding RNA. Genom. Proteom. Bioinform. 15, 177–186 (2017).
Beermann, J., Piccoli, M. T., Viereck, J. & Thum, T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol. Rev. 96, 1297–1325 (2016).
Li, M. et al. Long non-coding RNAs in renal cell carcinoma: a systematic review and clinical implications. Oncotarget 8, 48424–48435 (2017).
Ulrich., H., Weidle, Fabian, B., Gwen, K. & Rüdiger, R. Long non-coding RNAs and their role in metastasis. Cancer Genom. Proteom. 14, 143–160 (2017).
Li, J., Tian, H., Yang, J. & Gong, Z. Long noncoding RNAs regulate cell growth, proliferation, and apoptosis. DNA Cell Biol. 35, 459–470 (2016).
Montes, M. & Lund, A. H. Emerging roles of lncRNAs in senescence. FEBS J. 283, 2414–2426 (2016).
Wu, H. et al. Genetic variants of lncRNA HOTAIR and risk of epithelial ovarian cancer among Chinese women. Oncotarget 7, 41047–41052 (2016).
Zhong, X., Hu, X. & Zhang, L. Oncogenic long noncoding RNA FAL1 in human cancer. Mol. Cell. Oncol. 2, e977154 (2015).
Zhong, Y., Gao, D., He, S., Shuai, C. & Peng, S. Dysregulated expression of long noncoding RNAs in ovarian cancer. Int. J. Gynecol. Cancer 26, 1564–1570 (2016).
Xia, B. et al. Long non-coding RNA ZFAS1 interacts with miR-150-5p to regulate Sp1 expression and ovarian cancer cell malignancy. Oncotarget 8, 19534–19546 (2017).
Özeş, A. R. et al. Therapeutic targeting using tumor specific peptides inhibits long non-coding RNA HOTAIR activity in ovarian and breast cancer. Sci. Rep. 7, 894 (2017).
Hu, X. et al. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell 26, 344–357 (2014).
Liu, R. T. et al. Effects of lncRNA-HOST2 on cell proliferation, migration, invasion, and apoptosis of human hepatocellular carcinoma cell line SMMC-7721. Biosci. Rep. 37, BSR20160532 (2017).
Huang, L. et al. Role of Wnt/β-catenin, Wnt/c-Jun N-terminal kinase and Wnt/Ca(2+) pathways in cisplatin-induced chemoresistance in ovarian cancer. Exp. Ther. Med. 12, 3851–3858 (2016).
Cheng, Z. et al. A long noncoding RNA AB073614 promotes tumorigenesis and predicts poor prognosis in ovarian cancer. Oncotarget 6, 25381–25389 (2015).
Rinn, J. L. & Chang, H. Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166 (2012).
Cesana, M. et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147, 358–369 (2011).
Wang, H. et al. LncRNAs expression profiling in normal ovary, benign ovarian cyst, and malignant epithelial ovarian cancer. Sci. Rep. 6, 38983 (2016).
Wu, T. & Du, Y. LncRNAs: from basic research to medical application. Int J. Biol. Sci. 13, 295–307 (2017).
Kladi-Skandali, A., Michaelidou, K., Scorilas, A. & Mavridis, K. Long noncoding RNAs in digestive system malignancies: a novel class of cancer biomarkers and therapeutic targets? Gastroenterol. Res. Pract. 2015, 319861 (2015).
Wang, J. et al. Downregulated lncRNA HOTAIR expression in ovarian cancer stem cells decreases its tumorgeniesis and metastasis by inhibiting epithelial-mesenchymal transition. Cancer Cell Int. 15, 24 (2015).
Qiu, J. J. et al. The long non-coding RNA ANRIL promotes proliferation and cell cycle progression and inhibits apoptosis and senescence in epithelial ovarian cancer. Oncotarget 7, 32478–32492 (2016).
Ren, C. et al. Functions and mechanisms of long noncoding RNAs in ovarian cancer. Int. J. Gynecol. Cancer 25, 566–569 (2015).
Chaudhary, R. et al. Prosurvival long noncoding RNA PINCR regulates a subset of p53 targets in human colorectal cancer cells by binding to Matrin 3. Elife 6, e23244 (2017).
Wang, B. et al. Increased MIR31HG lncRNA expression increases gefitinib resistance in non-small cell lung cancer cell lines through the EGFR/PI3K/AKT signaling pathway. Oncol. Lett. 13, 3494–3500 (2017).
Li, H., Jiang, X. & Niu, X. Long non-coding RNA reprogramming (ROR) promotes cell proliferation in colorectal cancer via affecting p53. Med. Sci. Monit. 23, 919–928 (2017).
Wang, Q. et al. Upregulated NNT-AS1, a long noncoding RNA, contributes to proliferation and migration of colorectal cancer cells in vitro and in vivo. Oncotarget 8, 3441–3453 (2017).
Wang, D., Wang, D., Wang, N., Long, Z. & Ren, X. Long non-coding RNA BANCR promotes endometrial cancer cell proliferation and invasion by regulating MMP2 and MMP1 via ERK/MAPK signaling pathway. Cell Physiol. Biochem. 40, 644–656 (2016).
Zhao, H. et al. Long noncoding RNA HEIH promotes melanoma cell proliferation, migration and invasion via inhibition of miR-200b/a/429. Biosci. Rep. 37, BSR20170682 (2017).
Marín-Béjar, O. et al. The human lncRNA LINC-PINT inhibits tumor cell invasion through a highly conserved sequence element. Genome Biol. 18, 202 (2017).
Yang, W., Li, Y., Song, X., Xu, J. & Xie, J. Genome-wide analysis of long noncoding RNA and mRNA co-expression profile in intrahepatic cholangiocarcinoma tissue by RNA sequencing. Oncotarget 8, 26591–26599 (2017).
Acknowledgements
This work was supported by the Foundation of National Natural Science Youth Foundation of China (No. 81401502), the Natural Science Foundation of Heilongjiang Province of China (No. H2018020), and the Postdoctoral Science Foundation of Heilongjiang Province of China (No. LBH-Q17120). The funders had no role in study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by I. Harris
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gao, C., Zhao, D., Zhao, Q. et al. Microarray profiling and co-expression network analysis of lncRNAs and mRNAs in ovarian cancer. Cell Death Discov. 5, 93 (2019). https://doi.org/10.1038/s41420-019-0173-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41420-019-0173-7
This article is cited by
-
The pivotal role of long non-coding RNAs as potential biomarkers and modulators of chemoresistance in ovarian cancer (OC)
Human Genetics (2024)
-
Long non-coding RNA HEIH: a novel tumor activator in multiple cancers
Cancer Cell International (2021)
-
Commensal microbes and p53 in cancer progression
Biology Direct (2020)
-
Novel role of lncRNA CHRF in cisplatin resistance of ovarian cancer is mediated by miR-10b induced EMT and STAT3 signaling
Scientific Reports (2020)