Abstract
Cetuximab is approved for the treatment of metastatic colorectal cancer (mCRC) with RAS wild-type. Nevertheless, the prognosis remains poor and the effectiveness of cetuximab is limited in KRAS mutant mCRC. Recently, emerging evidence has shown that ferroptosis, a newly discovered form of nonapoptotic cell death, is closely related to KRAS mutant cells. Here, we further investigated whether cetuximab-mediated regulation of p38/Nrf2/HO-1 promotes RSL3-induced ferroptosis and plays a pivotal role in overcoming drug resistance in KRAS mutant colorectal cancer (CRC). In our research, we used two KRAS mutant CRC cell lines, HCT116 and DLD-1, as models of intrinsic resistance to cetuximab. The viability of cells treated with the combination of RSL3 and cetuximab was assessed by the CCK-8 and colony formation assays. The effective of cetuximab to promote RSL3-induced ferroptosis was investigated by evaluating lipid reactive oxygen species accumulation and the expression of the malondialdehyde and the intracellular iron assay. Cetuximab therapy contributed to regulating the p38/Nrf2/HO-1 axis, as determined by western blotting and transfection with small interfering RNAs. Cetuximab promoted RSL3-induced ferroptosis by inhibiting the Nrf2/HO-1 in KRAS mutant CRC cells, and this was further demonstrated in a xenograft nude mouse model. Our work reveals that cetuximab enhances the cytotoxic effect of RSL3 on KRAS mutant CRC cells and that cetuximab enhances RSL3-induced ferroptosis by inhibiting the Nrf2/HO-1 axis through the activation of p38 MAPK.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) is the third most deadly cancer in the world [1, 2]. Despite the development of diagnostic and therapeutic strategies, CRC remains one of the most severe human cancers due to resistance to treatment.
In recent years, great progress has been made in the combination of cetuximab and chemotherapy in the treatment of metastatic colorectal cancer (mCRC) [3]. However, KRAS mutations are seen in 36–46% of CRC cases, and they do not benefit from cetuximab treatment [4, 5]. Treatment with cetuximab is ineffective in CRC patients with KRAS mutations [6, 7]. Therefore, we considered how to develop the therapeutic efficacy of cetuximab in KRAS mutant CRC in our study.
Ferroptosis is a newly discovered form of nonapoptotic cell death that is triggered via iron-dependent lipid peroxidation [8]. The mechanisms of ferroptosis are distinct from those of typical cell death processes, such as necrosis, autophagy, and apoptosis [9]. RAS-selective lethal 3 (RSL3), a small molecule, can kill RAS mutant cancer cells [10] and activate iron-dependent, nonapoptotic cell death of RAS mutant cancer cells [11]. RSL3 is a potent ferroptosis-triggering agent that directly inhibits GPX4, which can promote ferroptosis, including CRC cells [12]. Thus, there might be special links between ferroptosis and KRAS mutant CRC.
Nuclear factor erythroid 2-related factor 2 (Nrf2), a dominant regulator of antioxidant transcription factors, prevents lipid peroxidation [13] and ferroptosis by increasing the transcription of multiple cytoprotective enzymes, such as haem oxygenase-1 (HO-1) [14]. Ferroptosis can be alleviated potently via Nrf2/HO-1 axis activation through the abolishment of lipid oxidation [15, 16]. Therefore, suppressing Nrf2/HO-1 signalling could be a potential target in promoting ferroptosis. p38 mitogen-activated protein kinase (p38 MAPK) can be activated by cetuximab [17], and p38 MAPK has been confirmed to be involved in the regulation of Nrf2/HO-1 [18, 19]. Accordingly, we speculated that activation of p38 MAPK plays a major role in suppressing Nrf2/HO-1 signalling.
Here, our study showed that combined treatment with RSL3 and cetuximab increases the death of HCT116 and DLD-1 KRAS mutant CRC cell lines, leading to the accumulation of lipid peroxide products and decreased cell viability. Significantly, we for the first time demonstrated that cetuximab can promote RSL3-induced ferroptosis by markedly suppressing Nrf2/HO-1 signalling via p38 MAPK activation in KRAS mutant CRC cell lines.
Results
Cetuximab enhances the cytotoxicity effect of RSL3 treatment on KRAS mutant CRC cells
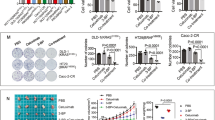
Four KRAS mutant CRC cell lines (HCT116, DLD-1, LOVO and SW480) were treated in indicated concentration of RSL3 alone or in combination with cetuximab (100 μg/ml) for 24 h. (Fig. S1). We found that combination treatment with RSL3 and cetuximab enhanced the death of HCT116 and DLD-1 cells compared with RSL3 alone. In subsequent experiments, HCT116 and DLD-1 cells were chosen and treated with RSL3 (1 μM), cetuximab (100 μg/ml) for 24 h. We found that treatment with cetuximab enhanced the cytotoxic effect of RSL3 in HCT116 and DLD-1 cells (Fig. 1A, B).
A HCT116 and DLD-1 cells were treated with RSL3 (1 μM), cetuximab (100 μg/ml) or their combination for 24 h. The inhibitory effects were determined by the CCK-8 assay. B Cell morphology changes were observed by inverted light microscopy. Scale bar, 100 μm. C The colony formation with the indicated treatment was performed (left panel), and a histogram of colony numbers is shown (right panel). **P < 0.01.
We next investigated whether cetuximab enhances antiproliferative effects on RSL3-treated HCT116 and DLD-1 cells, and the colony formation assay was performed. Compared with RSL3 or cetuximab alone, combination treatment with RSL3 and cetuximab significantly suppressed proliferation (Fig. 1C). All these results suggest that cetuximab enhances the cytotoxic effect of RSL3 on KRAS mutant CRC cell lines by significantly inhibiting cell viability and inhibiting cell proliferation.
Cetuximab enhances RSL3-induced ferroptosis in KRAS mutant CRC cells
RSL3 is a potent ferroptosis activator [20]. To investigate whether ferroptosis was further promoted by treatment with RSL3 and cetuximab, two indicators of ferroptosis, i.e., lipid reactive oxygen species (ROS) accumulation and malondialdehyde (MDA) levels, were assessed in KRAS mutant HCT116 and DLD-1 cells. Then, C11‐BODIPY581/591, a lipid peroxidation probe, was applied to measure the level of intracellular lipid ROS. Consistent with our expectation, following co-treatment with RSL3 and cetuximab, lipid ROS accumulation was remarkably increased (Fig. 2A). Moreover, MDA levels were remarkably increased after co-treatment with RSL3 and cetuximab in HCT116 and DLD-1 cells (Fig. 2B). Intriguingly, treatment with cetuximab did not affect lipid ROS accumulation or MDA levels in either HCT116 or DLD-1 cell lines but enhanced the RSL3-induced increases in lipid ROS levels and MDA levels. These results indicate that ferroptosis might be promoted after co-treatment with RSL3 and cetuximab in KRAS mutant CRC cell lines. Ferroptosis is characterized by iron-dependent accumulation [9]. Thus, intracellular iron levels were determined in HCT116 and DLD-1 cells by using a Fe2+ iron probe known as FerroOrange. The cells co-treated with RSL3 and cetuximab emitted much stronger orange fluorescence compared with treated with RSL3 alone (Fig. 2C).
A HCT116 and DLD-1 cells were treated with RSL3 (1 μM), cetuximab (100 μg/ml) or their combination for 24 h. The cellular lipid ROS level was analyzed by a flow cytometer. B HCT116 and DLD-1 cells were treated with RSL3 (1 μM), cetuximab (100 μg/ml) or their combination for 24 h. Intracellular MDA levels were measured by an MDA Assay Kit. C HCT116 and DLD-1 cells were treated with RSL3 (1 μM), cetuximab (100 μg/ml) or their combination for 24 h. Chelatable iron levels were determined using FerroOrange. Scale bar, 50 μm. D HCT116 and DLD-1 cells were treated with cetuximab (100 μg/ml) or RSL3 (1 μM) in the absence or presence of Fer-1 (1 μM) for 24 h, and cell viability was evaluated by the CCK-8 assay. E HCT116 and DLD-1 cells were treated with cetuximab (100 μg/ml) or RSL3 (1 μM) in combination with Z-VAD-FMK (10 μM), necrostatin-1 (10 μM) or 3-MA (60 μM) for 24 h, and cell viability was assessed by the CCK-8 assay. **P < 0.01; *P < 0.05; NS: no significance.
Moreover, treatment with RSL3 and cetuximab increased the inhibition of HCT116 and DLD-1 cell growth, and this effect was reversed by the ferroptosis rescue agent Fer-1 (Fig. 2D) but not by the rescue agent of apoptosis (Z-VAD-FMK), necroptosis (necrostatin-1), or autophagy (3-MA) (Fig. 2E). Collectively, these findings strongly suggest that cetuximab enhances RSL3-induced ferroptosis in KRAS mutant CRC cell lines.
Combination treatment with RSL3 and cetuximab inhibits Nrf2/HO-1 axis in KRAS mutant CRC cells
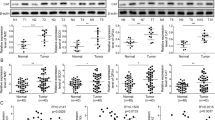
Kelch-like ECH-associated protein-1 (Keap1) is a master regulator in cellular responding to oxidative stress. Following oxidative stress, Keap1 is dissociated from Nrf2, leading to Nrf2 accumulation, nuclear translocation, and activate the transcription of target gene including HO-1 [21]. Our study found that following treatment with RSL3 and cetuximab increasing Keap1 expression, and inhibiting Nrf2/HO-1 signalling, as shown by western blot analysis (Fig. 3A).
A The protein levels of Keap1, Nrf2 and HO-1 in HCT116 and DLD-1 cells were measured by western blotting after treatment with RSL3 (1 μM), cetuximab (100 μg/ml) or their combination for 24 h. B Knockdown of Nrf2 by siRNA reduced the expression of Nrf2 and the protein levels of Nrf2 and HO-1 after treatment with RSL3 (1 μM) for 24 h. C siRNA-mediated knockdown of Nrf2 increased the sensitivity of HCT116 and DLD-1 cells to RSL3. D–F The levels of MDA, lipid ROS and intracellular iron were measured in Nrf2-silenced HCT116 and DLD-1 cells pretreated with or without RSL3 (1 μM). Scale bar, 100 μm. **P < 0.01; *P < 0.05.
To evaluate the effects of the expression of Nrf2 on KRAS mutant CRC cells treated with cetuximab and RSL3, we designed two small interfering RNAs (siRNAs) targeting Nrf2. In HCT116 and DLD-1 cells, siRNA transfection for 48 h period reduced the expression of Nrf2 and its downstream target HO-1. Compared with the RSL3 group, the Nrf2 and HO-1 level in the RSL3-treated transfected groups was reduced (Fig. 3B). Transfection of an siRNA targeting Nrf2 increased the sensitivity of HCT116 and DLD-1 cells to RSL3 (Fig. 3C).
We sought to further confirm whether knockdown of Nrf2 can cause lipid oxidation, and we measured the levels of MDA and intracellular ROS in Nrf2-siRNA-transfected HCT116 and DLD-1 cells. As expected, knockdown of Nrf2 enhanced RSL3-induced MDA levels (Fig. 3D). Then, we used flow cytometry to analyze the intracellular lipid ROS level, and the results demonstrated that knockdown of Nrf2 promoted RSL3-induced oxidation of lipids in HCT116 and DLD-1 cells (Fig. 3E). Additionally, the results showed that knockdown of Nrf2 enhanced RSL3-induced intracellular iron accumulation (Fig. 3F). These results indicate that combination treatment with RSL3 and cetuximab inhibits Nrf2/HO-1 signalling in KRAS mutant CRC cells.
Cetuximab activates p38 MAPK and regulates the Nrf2/HO-1 axis
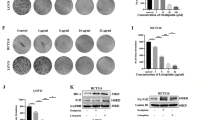
p38 MAPK has been reported to regulate antioxidant enzymes expression, including Nrf2 and HO-1 [22]. To investigate the role of p38 MAPK in the expression of Nrf2/HO-1, we evaluated the effect of cetuximab and SB202190 (specific inhibitors of p38) on p38 activity in HCT116 and DLD-1 cells. The protein level of p38 in cetuximab- or SB202190-treated cells was not different from that in control. The levels of p-p38 were significantly enhanced by cetuximab treatment, but this effect was abolished when co-treatment with cetuximab and SB202190 (Fig. 4A). We then investigated whether inhibition of p38 expression can affect cetuximab-induced inhibition of Nrf2 and HO-1 expression. Western blot analysis demonstrated that treatment with cetuximab decreased the expression of Nrf2 and HO-1, and SB202190 reduced cetuximab-mediated inhibition of Nrf2 and HO-1 (Fig. 4A). Moreover, the death of HCT116 and DLD-1 cells induced by treatment with RSL3 and cetuximab was partly rescued by SB202190 (Fig. 4B). To further verify if Nrf2/HO-1 pathway is involved in which cetuximab sensitizes cells to RSL3, HCT116 and DLD-1 cells were transient transfected with Nrf2 or HO-1 expressing vectors. As shown in Fig. 4C, D, overexpression of Nrf2 restored the combination treatment with cetuximab and RSL3-induced inhibition of HO-1 expression and cell growth in HCT116 and DLD-1 cells. Also, overexpression of HO-1 reversed the growth inhibition induced by cetuximab and RSL3 combination treatment in CRC cells (Fig. 4E, F). In addition, activation of Nrf2 by t-BHQ treatment or activation of HO-1 by hemin treatment partly reversed the growth inhibition of HCT116 and DLD-1 cells induced by RSL3 and cetuximab treatment (Fig. 4G, H). In addition, knockdown of Keap1 significantly restored theNrf2 and HO-1, and reduced the cytotoxicity induced by co-treatment cetuximab and RSL3 in HCT116 and DLD-1 cells (Fig. S2). These results demonstrate that p38 MAPK participates in the regulation of Nrf2 and HO-1 expression and that cetuximab can inhibit the Nrf2/HO-1 pathway via p38 MAPK activation.
A Western blot analysis of p-p38, total p38, Nrf2 and HO-1 expression levels in HCT116 and DLD-1 cells incubated with cetuximab (100 μg/ml), SB202190 (1 μM) or cetuximab in combination with SB202190 for 24 h. B HCT116 and DLD-1 cells were treated with cetuximab (100 μg/ml) or RSL3 (1 μM) in the absence or presence of SB202190 (1 μM) for 24 h, and cell viability was assessed by the CCK-8 assay. C The protein levels of Nrf2 and HO-1 in HCT116 and DLD-1 cells or Nrf2 overexpressed HCT116 and DLD-1 cells treated with RSL3 (1 μM) combination with cetuximab (100 μg/ml) for 24 h. D HCT116 and DLD-1 cells with overexpression Nrf2 were treated with or without RSL3 (1 μM) combination with cetuximab (100 μg/ml) for 24 h. Cell viability was assessed by CCK-8 assays. E The protein level of HO-1 in HCT116 and DLD-1 cells or HO-1 overexpressed HCT116 and DLD-1 cells treated with RSL3 (1 μM) combination with cetuximab (100 μg/ml) for 24 h. F HCT116 and DLD-1 cells with overexpression HO-1 were treated with or without RSL3 (1 μM) combination with cetuximab (100 μg/ml) for 24 h. Cell viability was assessed by CCK-8 assays. G–H HCT116 and DLD-1 cells were treated with cetuximab (100 μg/ml) or RSL3 (1 μM) with or without t-BHQ (20 μM) or hemin (20 μM) for 24 h, and cell viability was assessed by the CCK-8 assay. **P < 0.01.
Cetuximab enhances RSL3-induced ferroptosis by suppressing the Nrf2/HO-1 axis in KRAS mutant CRC cells in a xenograft nude mouse model
To further explored whether cetuximab promotes RSL3-induced ferroptosis in vivo, a DLD-1 xenograft nude mouse model was established. A schematic of tumour inoculation and systemic injection is shown in Fig. 5A. All the mice survived well after cell implantation or were treated with vehicle, RSL3, cetuximab, or RSL3 in combination with cetuximab. Following treatment for 17 days, the size of the tumour gradually increased in the control and cetuximab treatment group. However, administration of RSL3 alone and treatment with both agents led to a decrease in tumour size, and the in the both agents’ administration group exhibited the greatest reduction in tumours size (Fig. 5B, C) and tumour volume (Fig. 5D). Body weight and daily food intake did not change significantly in the control or treatment groups (Fig. 5E). We then tested whether combinatory treatment with cetuximab and RSL3 can regulate lipid peroxidation in vivo. Strikingly, MDA concentration levels were significantly enhanced in the combined group (Fig. 5F).
A Schematic of the anticancer effect of the combination of RSL3 (5 mg/kg) and cetuximab (13 mg/kg) in a DLD-1 cell subcutaneous tumour model. B The tumour weights of excised tumours on day 19. C Excised tumours on day 19. D The tumour volume of each group was calculated every 3 days. E The body weight of each group was calculated every 3 days. F The level of MDA in excised tumours on day 19 was assayed. **P < 0.01.
Next, we measured Nrf2, HO-1 and Keap1 protein levels. The western blotting date showed that the relative levels of Nrf2 and HO-1 were decreased and that keap1 expressed was relatively increased after co-treatment with RSL3 and cetuximab, as was observed in vitro (Fig. 6A). Then, we measured the ki67, Keap1, Nrf2, and HO-1 expression by immunohistochemical staining. Consistent with the western blotting results, the immunohistochemical analysis showed that low expression of Ki67, Nrf2, and HO-1 and high expression of keap1 were found after co-treatment with RSL3 and cetuximab (Fig. 6B). Taken together, these in vivo results further support the in vitro evidence that cetuximab enhances RSL3-induced ferroptosis by suppressing Nrf2/HO-1 activation.
Discussion
CRC is the most common primary malignancy of the colorectum and progresses rapidly [23]. Moreover, KRAS is the most common form of mutated oncogene in cancer, and the frequency of KRAS mutations is particularly high in colorectal, pancreatic, and lung cancers [24,25,26]. Cetuximab, an EGFR-targeting agent, is a standard treatment for KRAS wild-type mCRC [27]. KRAS mutant CRC cells have been reported that it could promote tumour progression and induce resistance to cetuximab therapy [28]; however, there is no effective treatment to specifically treat cancers expressing KRAS mutations [29, 30].
Despite the failure of cetuximab in treating KRAS mutant CRC, development efforts and substantial research in this area have continued. Suto et al. confirmed that miR-7 regulates the sensitive of cetuximab by inhibiting EGFR signalling in KRAS mutant CRC cells [31]. Napolitano [32] and Troiani [33] et al. showed that co-treatment with cetuximab and regorafenib induced synergistic antiproliferative and apoptotic effects to overcome resistance to cetuximab by inhibiting MAPK and AKT pathways. Han et al. demonstrated that high expression of MLH1 increase cetuximab sensitivity by blocking HER-2 signalling [34].
Recently, research has shown that activation of ferroptosis effectively prevents tumour progression and enhances the efficacy of targeted therapy and chemotherapy. For example, it has been reported that vitamin C can lead to ferroptosis, and co-treatment with vitamin C and cetuximab could restrain the emergence of acquired resistance to cetuximab in RAS/BRAF wild-type CRC [35]. Upregulation of MT-1G expression protects hepatocellular carcinoma cells from the effect of sorafenib and promotes cancer progression by inhibiting ferroptosis [36]. Short-treatment of erastin significantly increases the cytotoxic effects of cisplatin [37]. The combination of a low concentration of paclitaxel and RSL3 induces ferroptosis in mutant p53 hypopharyngeal squamous cell carcinoma [38]. The combination of β-elemene, a bioactive compound, and cetuximab is effective to KRAS mutant CRC cells by inducing ferroptosis [39]. Additionally, in our study, we found that the cetuximab enhanced the cytotoxic effect of RSL3 on KRAS mutant CRC cells. Ferroptosis agent, but not the inhibitor of caspase, autophagy, or necroptosis, rescued the cell death induced by co-treatment with RSL3 and cetuximab, indicating that in the presence of cetuximab, ferroptosis is a key process that contributes to cell death. Interestingly, we found that cetuximab promoted RSL3-induced ferroptosis in KRAS mutant CRC cells.
Mechanistically, we demonstrated that Nrf2/HO-1 is essential for cetuximab to promote RSL3-induced ferroptosis in KRAS mutant CRC cells. Disruption of the Nrf2/HO-1 axis was previously shown to contribute to the promotion of ferroptosis [40,41,42]. In this study, we assessed the expression of Nrf2/HO-1 both in vitro and in vivo. The administration of cetuximab to RSL3-treated cells further promoted ferroptosis by decreasing Nrf2 and HO-1 expression. We determined that treatment with cetuximab significantly enhanced the cytotoxicity of RSL3 in KRAS-mutated CRC cells and promoted RSL3-induced ferroptosis by suppressing Nrf2/HO-1 expression. These results indicated that cetuximab decreased the activation of Nrf2/HO-1 in RSL3-treated cells, which could in turn increased RSL3-induced lipid ROS and MDA levels. In addition, the p38 MAPK pathway was evaluated in this study. p38 MAPK is the upstream molecule of Nrf2 [43, 44]. Previous studies have shown that nitric oxide induces HO-1 expression in HeLa cells [45] and that andrographolide induces Nrf2 and HO-1 expression in astrocytes [46], partly via p38 MAPK. In addition, p38 MAPK plays an important role in regulating the expression of Nrf2/HO-1 in brain astrocytes [47]. Ferroptosis has a broad clinical application prospect in cancer therapy [9]. In our study, we found that cetuximab combined with ferroptosis inducer treatment was effective in KRAS mutations CRC. Future preclinical studies are remine necessary to test whether ferroptosis-triggering agent combined with cetuximab could intensify the efficacy in KRAS mutant CRC.
In conclusion, our study revealed that cetuximab enhances the cytotoxic effect of RSL3 on KRAS mutant CRC cells, and we demonstrated a novel mechanism involved in the anticancer effect of cetuximab on KRAS mutant CRC. Cetuximab enhanced RSL3-induced ferroptosis by inhibiting the Nrf2/HO-1 axis through the activation of p38 MAPK (Fig. 7). Our data suggests that it has the potential to be translated into clinical trials testing the effects of cetuximab combined treatment with ferroptosis inducer in KRAS mutant CRC patients. This finding will hopefully help in the development of an attractive treatment strategy for KRAS mutant CRC patients.
Materials and methods
Cell lines
Human colorectal cancer cell lines HCT116, DLD-1, LOVO and SW480 were obtained from the Chinese Academy of Sciences (Shanghai, China). HCT116 cells were cultured in DMED (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% foetal bovine serum (Thermo Fisher Scientific). DLD-1, LOVO and SW480 cells were cultured in RPMI 1640 medium (Thermo Fisher Scientific) supplemented with 10% FBS. The cells were maintained in a 5% CO2 humidified incubator at 37 °C.
Reagents and antibodies
Cetuximab was purchased from Merck (Darmstadt, Germany), and RSL3 (HY-100218A) and SB202190 (HY-10295) were purchased from MedChemExpress (Monmouth Junction, NJ, USA). Necrostatin-1 (GC11008) and Z-VAD-FMK (GC12861) were purchased from GLPBIO (Montclair, CA, USA). Ferrostatin-1 (Fer-1) (HY-100579) and 3-MA (HY-19312) were purchased from MedChemExpress. Primary antibodies against Keap1 (AF5266), HO-1 (AF5393), p38 (BF8015), and phosphorylated (p)-P38 (AF4001) were purchased from Affinity Biosciences (OH, USA). A Nrf2 antibody (16396-1-AP) was purchased from Proteintech (Rosemont, IL, USA). A Ki67 antibody (ab15580) was purchased from Abcam (Cambridge, United Kingdom).
Cell viability assay
The viability of CRC cells was determined by a Cell Counting Kit 8 (CCK-8, Dojindo, Japan). In brief, cells (3000-5000 per well) were plated in 96-well plates. Then, 100 μl of medium was added to cells containing 10 μl of CCK-8 solutions incubated at 37 °C for 3 h. Absorbance was measured at 450 nm wavelength.
Western blot analysis
Protein concentrations were determined using a BCA Protein Assay Kit (Thermo Fisher Scientific). Proteins (30 μg) were electrophoresed on 12% SDS-PAGE gels and were transferred to PVDF membranes, which were blocked in 5% BSA for 2 h and were incubated with primary antibodies at 4 °C overnight. Then, carefully transferred to secondary antibody for 1 h. Next, we used TBST solution to wash the membranes three times. Antibody signals were detected using an ECL detection system (Bio-Rad, California, USA).
Colony formation assays
To assess the colony formation of monolayer cultures, cells (1000 per well) were seeded into a 6-well plate. After two weeks, colonies were fixed and stained with 4% paraformaldehyde and 0.1% crystal violet for 30 min at room temperature.
Lipid reactive oxygen species (ROS) level assay
Lipid ROS was detected by C11-BODIPY® 581/591 (D3861, Thermo Fisher Scientific) [48]. After 24 h, C11-BODIPY was incubated in cells with medium at 37 °C for 30 min. The the results were analyzed with FlowJo V10 software [39].
Malondialdehyde (MDA) assay
MDA levels were measured with a Lipid Peroxidation MDA Assay Kit (#A003-4-1, Nanjing Jiancheng, China) according to the manufacturer’s instructions.
Intracellular iron assay
Intracellular iron levels were assessed by using FerroOrange (Dojindo, Japan) [49]. Cells were seeded in 6-well plates and treated with the indicated treatment. After 24 h, FerroOrange (1 mM) dispersed in serum-free medium was added to the cells, and the cells were incubated for 30 min at 37 °C. Finally, the fluorescence was detected by confocal microscope.
Small interfering RNA (siRNA) transfection
siRNA-Nrf2 or siRNA-Keap1 (GenePharma, Shanghai, China) were used to knocked down Nrf2 and Keap1. The sequences of the siRNAs: si-Nrf2-1 (sense: 5′-GCAUGCUACGUGAUG AAGATT-3′; antisense: 5′-UCUUCAUCACGUAGCAUG CTT-3′) and si-Nrf2-2 (sense: 5′-UCUUCAUCACGUAGCAUGCTT-3′; antisense: 5′-GCAUGCUAC GUGAUGAAGATT-3′), siKeap1-1 (sense: 5′-CCUCAAUCGUCUCCUUUAUTT-3′; antisense: 5′-AUAAAGGAGACGAUUGAGGTT-3′) and siKeap1-2 (sense: 5′-GUCCUGCA CAACUGUAUCUTT-3′; antisense: 5′-AGAUACAGUUGUGCAGGACTT-3′). Transfection was conducted with Lipofectamine 3000 (Thermo Fisher Scientific) according to the instructions of the manufacturer.
Transduction
The vector expressing Nrf2 or HO-1 gene was obtained from GenePharma. HCT116 and DLD-1 cells were transient transfected with Nrf2 or HO-1 expressing vector. The upregulated expression of Nrf2 or HO-1 was confirmed by western blotting.
Xenograft tumour models
The DLD-1 cell suspension (4 × 106 cells/200 μl) was injected subcutaneously into the right dorsal flank of 5-week-old male BALB/c nude mice (Charles River, China). The mice were randomly divided into four groups (5 mice/group): 1) the control group, 2) the RSL3 group, 3) the cetuximab group, and 4) the RSL3 + cetuximab group. Both RSL3 (5 mg/kg) and cetuximab (13 mg/kg) were administered by intraperitoneal injection in a volume of 100 μl once per day. The tumour volume was calculated as 0.5 × length × width2 [50]. After 17 days of treatment, the mice were sacrificed, and the tumours were removed. Then, tumour tissue obtained from the different treated groups was subjected to western blotting and immunohistochemical experiments. The experiment was approved by the First Affiliated Hospital of Wenzhou Medical University Experimental Animal Ethics Committee.
Immunohistochemistry (IHC)
Xenograft tumour tissues were fixed, dehydrated, embedded in paraffin, and sectioned (4 μm). The slides were incubated with the following primary antibodies: Ki67 (1:400), Keap1 (1:600), Nrf2 (1:50), and HO-1 (1:300).
Statistical analysis
Statistical analysis was performed with GraphPad Prism 7.0 software (San Diego, CA, USA). p < 0.05 and p < 0.01 were defined as statistically significant and highly significant, respectively, compared with the control or indicated group. p-values were analyzed by using Student’s t test or one-way ANOVA. All data were generated from three independent experiments.
Data availability
All data are included in this article and its supplementary materials or available upon request to the corresponding author.
References
Siegel R, Miller K, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30.
Ferlay J, Colombet M, Soerjomataram I, Parkin D, Piñeros M, Znaor A, et al. Cancer statistics for the year 2020: an overview. Int J Cancer. 2021;149:89–778.
Jensen B, Schou J, Yilmaz M, Johannesen H, Skougaard K, Linnemann D, et al. Cetuximab plus irinotecan administered biweekly with reduced infusion time to heavily pretreated patients with metastatic colorectal cancer and related RAS and BRAF mutation status. Int J Cancer. 2020;148:2542–56.
Manzanares-Martin B, Cebrián Aranda A, Del Puerto-Nevado L, González R, Solanes S, Gómez-España M, et al. Improving selection of patients with metastatic colorectal cancer to benefit from cetuximab based on KIR genotypes. J Immunother Cancer. 2021;9:e001705.
Serebriiskii I, Connelly C, Frampton G, Newberg J, Cooke M, Miller V, et al. Comprehensive characterization of RAS mutations in colon and rectal cancers in old and young patients. Nat Commun. 2019;10:3722.
Wadlow R, Hezel A, Abrams T, Blaszkowsky L, Fuchs C, Kulke M, et al. Panitumumab in patients with KRAS wild-type colorectal cancer after progression on cetuximab. Oncologist. 2012;17:14.
Amado R, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman D, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–34.
Hirschhorn T, Stockwell B. The development of the concept of ferroptosis. Free Radic Biol Med. 2019;133:130–43.
Dixon S, Lemberg K, Lamprecht M, Skouta R, Zaitsev E, Gleason C, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–72.
Yang W, Stockwell B. Synthetic lethal screening identifies compounds activating iron-dependent, nonapoptotic cell death in oncogenic-RAS-harboring cancer cells. Chem Biol. 2008;15:234–45.
Chen X, Kang R, Kroemer G, Tang D. Broadening horizons: the role of ferroptosis in cancer. Nat Rev Clin Oncol. 2021;18:280–96.
Shintoku R, Takigawa Y, Yamada K, Kubota C, Yoshimoto Y, Takeuchi T, et al. Lipoxygenase-mediated generation of lipid peroxides enhances ferroptosis induced by erastin and RSL3. Cancer Sci. 2017;108:2187–94.
Dodson M, Castro-Portuguez R, Zhang DD. NRF2 plays a critical role in mitigating lipid peroxidation and ferroptosis. Redox Biol. 2019;23:101107.
Liu X, Yuan X, Liang G, Zhang S, Zhang G, Qin Y, et al. BRG1 protects the heart from acute myocardial infarction by reducing oxidative damage through the activation of the NRF2/HO1 signaling pathway. Free Radic Biol Med. 2020;160:820–36.
Sun J, Zhou C, Zhao Y, Zhang X, Chen W, Zhou Q, et al. Quiescin sulfhydryl oxidase 1 promotes sorafenib-induced ferroptosis in hepatocellular carcinoma by driving EGFR endosomal trafficking and inhibiting NRF2 activation. Redox Biol. 2021;41:101942.
Qiu Y, Wan B, Liu G, Wu Y, Chen D, Lu M, et al. Nrf2 protects against seawater drowning-induced acute lung injury via inhibiting ferroptosis. Respir Res. 2020;21:232.
Marzi L, Combes E, Vié N, Ayrolles-Torro A, Tosi D, Desigaud D, et al. FOXO3a and the MAPK p38 are activated by cetuximab to induce cell death and inhibit cell proliferation and their expression predicts cetuximab efficacy in colorectal cancer. Br J Cancer. 2016;115:1223–33.
Yang C, Hsiao L, Lin H, Tseng H, Situmorang J, Leu Y, et al. Induction of HO-1 by 5, 8-dihydroxy-4’,7-dimethoxyflavone via activation of ROS/p38 MAPK/Nrf2 attenuates thrombin-induced connective tissue growth factor expression in human cardiac fibroblasts. Oxid Med Cell Longev. 2020;2020:1080168.
Chen Q, Shen L, Chen C, He H, Fu Y, Xu L, et al. Cetuximab combined with cisplatin improves the prognosis of gastric cancer patients and its effect on P38 MAPK expression. J BUON. 2019;24:2490–8.
Zhou B, Liu J, Kang R, Klionsky D, Kroemer G, Tang D. Ferroptosis is a type of autophagy-dependent cell death. Semin Cancer Biol. 2020;66:89–100.
Tian Y, Wu K, Liu Q, Han N, Zhang L, Chu Q, et al. Modification of platinum sensitivity by KEAP1/NRF2 signals in non-small cell lung cancer. J Hematol Oncol. 2016;9:83.
Yao P, Nussler A, Liu L, Hao L, Song F, Schirmeier A, et al. Quercetin protects human hepatocytes from ethanol-derived oxidative stress by inducing heme oxygenase-1 via the MAPK/Nrf2 pathways. J Hepatol. 2007;47:253–61.
Andrei P, Battuello P, Grasso G, Rovera E, Tesio N, Bardelli A. Integrated approaches for precision oncology in colorectal cancer: the more you know, the better. Semin Cancer Biol. 2021. https://doi.org/10.1016/j.semcancer.2021.04.007
Jung H, Oh Y, Na D, Min S, Kang J, Jang D, et al. CRISPR screens identify a novel combination treatment targeting BCL-X and WNT signaling for KRAS/BRAF-mutated colorectal cancers. Oncogene. 2021;40:3287–302.
Noordhof A, Damhuis R, Hendriks L, de Langen A, Timens W, Venmans B, et al. Prognostic impact of KRAS mutation status for patients with stage IV adenocarcinoma of the lung treated with first-line pembrolizumab monotherapy. Lung cancer. 2021;155:163–9.
Valencia K, Erice O, Kostyrko K, Hausmann S, Guruceaga E, Tathireddy A, et al. The Mir181ab1 cluster promotes KRAS-driven oncogenesis and progression in lung and pancreas. J Clin Invest. 2020;130:1879–95.
Khelwatty S, Puvanenthiran S, Essapen S, Bagwan I, Seddon A, Modjtahedi H. RASHER2 expression is predictive of survival in cetuximab treated patients with wild type metastatic colorectal cancer. Cancers. 2021;13:638.
Liu H, Liang Z, Zhou C, Zeng Z, Wang F, Hu T, et al. Mutant KRAS triggers functional reprogramming of tumor-associated macrophages in colorectal cancer. Signal Transduct Target Ther. 2021;6:144.
Huang J, Zang Q, Wen Y, Pan Z, Yao Z, Huang M, et al. Prognostic value of KRAS mutation in patients undergoing pulmonary metastasectomy for colorectal cancer: a systematic review and meta-analysis. Crit Rev Oncol Hematol. 2021;160:103308.
Malapelle U, Passiglia F, Cremolini C, Reale M, Pepe F, Pisapia P, et al. RAS as a positive predictive biomarker: focus on lung and colorectal cancer patients. Eur J Cancer. 2021;146:74–83.
Suto T, Yokobori T, Yajima R, Morita H, Fujii T, Yamaguchi S, et al. MicroRNA-7 expression in colorectal cancer is associated with poor prognosis and regulates cetuximab sensitivity via EGFR regulation. Carcinogenesis. 2015;36:338–45.
Napolitano S, Martini G, Rinaldi B, Martinelli E, Donniacuo M, Berrino L, et al. Primary and acquired resistance of colorectal cancer to anti-EGFR monoclonal antibody can be overcome by combined treatment of regorafenib with cetuximab. Clin Cancer Res. 2015;21:2975–83.
Troiani T, Napolitano S, Vitagliano D, Morgillo F, Capasso A, Sforza V, et al. Primary and acquired resistance of colorectal cancer cells to anti-EGFR antibodies converge on MEK/ERK pathway activation and can be overcome by combined MEK/EGFR inhibition. Clin Cancer Res. 2014;20:3775–86.
Han Y, Peng Y, Fu Y, Cai C, Guo C, Liu S, et al. MLH1 deficiency induces cetuximab resistance in colon cancer via Her-2/PI3K/AKT signaling. Adv Sci (Weinh). 2020;7:2000112.
Lorenzato A, Magrì A, Matafora V, Audrito V, Arcella P, Lazzari L, et al. Vitamin C restricts the emergence of acquired resistance to EGFR-targeted therapies in colorectal cancer. Cancers (Basel). 2020;12:685.
Sun X, Niu X, Chen R, He W, Chen D, Kang R, et al. Metallothionein-1G facilitates sorafenib resistance through inhibition of ferroptosis. Hepatology. 2016;64:488–500.
Sato M, Kusumi R, Hamashima S, Kobayashi S, Sasaki S, Komiyama Y, et al. The ferroptosis inducer erastin irreversibly inhibits system x- and synergizes with cisplatin to increase cisplatin’s cytotoxicity in cancer cells. Sci Rep. 2018;8:968.
Ye J, Jiang X, Dong Z, Hu S, Xiao M. Low-concentration PTX and RSL3 inhibits tumor cell growth synergistically by inducing ferroptosis in mutant p53 hypopharyngeal squamous carcinoma. Cancer Manag Res. 2019;11:9783–92.
Chen P, Li X, Zhang R, Liu S, Xiang Y, Zhang M, et al. Combinative treatment of β-elemene and cetuximab is sensitive to KRAS mutant colorectal cancer cells by inducing ferroptosis and inhibiting epithelial-mesenchymal transformation. Theranostics. 2020;10:5107–19.
Lou J, Zhao L, Huang Z, Chen X, Xu J, Tai W, et al. Ginkgetin derived from Ginkgo biloba leaves enhances the therapeutic effect of cisplatin via ferroptosis-mediated disruption of the Nrf2/HO-1 axis in EGFR wild-type non-small-cell lung cancer. Phytomedicine. 2021;80:153370.
Ma H, Wang X, Zhang W, Li H, Zhao W, Sun J, et al. Melatonin suppresses ferroptosis induced by high glucose via activation of the Nrf2/HO-1 signaling pathway in type 2 diabetic osteoporosis. Oxid Med Cell Longev. 2020;2020:9067610.
Yang C, Zhang Y, Lin S, Liu Y, Li W. Suppressing the KIF20A/NUAK1/Nrf2/GPX4 signaling pathway induces ferroptosis and enhances the sensitivity of colorectal cancer to oxaliplatin. Aging (Albany NY). 2021;13:13515–34.
Yang Y, Cai X, Yang J, Sun X, Hu C, Yan Z, et al. Chemoprevention of dietary digitoflavone on colitis-associated colon tumorigenesis through inducing Nrf2 signaling pathway and inhibition of inflammation. Mol Cancer. 2014;13:48.
Hourihan J, Moronetti Mazzeo L, Fernández-Cárdenas L, Blackwell T. Cysteine sulfenylation directs IRE-1 to activate the SKN-1/Nrf2 antioxidant response. Mol Cell. 2016;63:553–66.
Chen K, Maines M. Nitric oxide induces heme oxygenase-1 via mitogen-activated protein kinases ERK and p38. Cell Mol Biol (Noisy-le-Gd). 2000;46:609–17.
Wong S, Tan M, Wong P, Herr D, Lai M. Andrographolide induces Nrf2 and heme oxygenase 1 in astrocytes by activating p38 MAPK and ERK. J Neuroinflammation. 2016;13:251.
Jeong Y, Park J, Kim D, Kim H. Lonchocarpine Increases Nrf2/ARE-Mediated Antioxidant Enzyme Expression by Modulating AMPK and MAPK Signaling in Brain Astrocytes. Biomol Ther (Seoul). 2016;24:581–8.
Ye Z, Hu Q, Zhuo Q, Zhu Y, Fan G, Liu M, et al. Abrogation of ARF6 promotes RSL3-induced ferroptosis and mitigates gemcitabine resistance in pancreatic cancer cells. Am J Cancer Res. 2020;10:1182–93.
Guo S, Yao X, Jiang Q, Wang K, Zhang Y, Peng H, et al. Dihydroartemisinin-loaded magnetic nanoparticles for enhanced chemodynamic therapy. Front Pharm. 2020;11:226.
Li B, Qi Z, He D, Chen Z, Liu J, Wong M, et al. NLRP7 deubiquitination by USP10 promotes tumor progression and tumor-associated macrophage polarization in colorectal cancer. J Exp Clin Cancer Res. 2021;40:126.
Acknowledgements
The authors would like to acknowledge the reviewers for their helpful comments on this paper.
Funding
This work is supported by the National Natural Science Foundation of China (No. 81572291) and Zhejiang Provincial Natural Science Foundation of China (No. LY19H160022 and No. Y22H162686) and Medicine and Health Technology Program of Zhejiang Province (No. 2021KY790).
Author information
Authors and Affiliations
Contributions
L.Y. and L.J. are the corresponding authors and they performed study design; J.Y. and J.M. performed the experiments; C.Y., W.C., X.Z., analyzed the data; J.D. provided technical support; J.M. revised the manuscript carefully; All authors read and approved the final paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics statement
This study was approved and consented by the ethics committee of the First Affiliated Hospital of Wenzhou Medical University.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by Boris Zhivotovsky
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yang, J., Mo, J., Dai, J. et al. Cetuximab promotes RSL3-induced ferroptosis by suppressing the Nrf2/HO-1 signalling pathway in KRAS mutant colorectal cancer. Cell Death Dis 12, 1079 (2021). https://doi.org/10.1038/s41419-021-04367-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-021-04367-3
This article is cited by
-
Synergism of non-thermal plasma and low concentration RSL3 triggers ferroptosis via promoting xCT lysosomal degradation through ROS/AMPK/mTOR axis in lung cancer cells
Cell Communication and Signaling (2024)
-
Crosstalk between metabolism and cell death in tumorigenesis
Molecular Cancer (2024)
-
Juglone induces ferroptosis in glioblastoma cells by inhibiting the Nrf2-GPX4 axis through the phosphorylation of p38MAPK
Chinese Medicine (2024)
-
Hypoxia, oxidative stress, and the interplay of HIFs and NRF2 signaling in cancer
Experimental & Molecular Medicine (2024)
-
Nrf2/HO-1 Alleviates Disulfiram/Copper-Induced Ferroptosis in Oral Squamous Cell Carcinoma
Biochemical Genetics (2024)