Abstract
Chemotherapy remains the primary treatment of advanced solid cancer, including lung cancer. However, as first-line treatment, cisplatin-based therapy is restricted by the frequent development of drug resistance. Increasing data showed that the programmed cell death protein ligand 1 (PD-L1) plays a vital role in regulating cisplatin resistance. However, the underlying mechanisms are not fully understood. We found that miR-526b-3p expression declined while PD-L1 was elevated in cisplatin-resistant lung cancer compared to that in cisplatin-sensitive lung cancer by analyzing clinical samples. Significantly, miR-526b-3p was associated with response to cisplatin negatively. We further demonstrated that miR-526b-3p reversed cisplatin resistance, suppressed metastasis, and activated CD8+ T cells in a STAT3/PD-L1-dependent manner. Thus, our findings extended the knowledge of PD-L1-mediated cisplatin resistance of lung cancer. In addition, the introduction of miR-526b-3p provided a new clue to improve the anti-tumor effects of the combination of immunotherapy and chemotherapy.
Similar content being viewed by others
Introduction
Platinum-based therapy is one of the most common chemotherapies against solid cancer, including lung cancer. Cisplatin, along with radiation therapy, is used to treat advanced lung cancer. Side effects and drug resistance are two main challenges that restrain the application and response of cisplatin [1]. Accumulating evidence shows that STAT3, MEK1, and AKT are frequently dysregulated in cisplatin-resistant cancer [2,3,4]. Therefore, therapies that targeted hyperactive genes are developed to diminish cisplatin resistance. However, the clinic outcomes of cisplatin-resistant cancer are not satisfactory.
miRNAs are endogenous noncoding RNA molecules that contain less than 25 nucleotides. miRNAs bind to the 3′-untranslated region (3′UTR) of target mRNAs, leading to mRNAs degradation and suppression. Therefore, miRNAs work as oncogenes and tumor suppressors in tumor malignancy. Nowadays, thousands of miRNAs molecules and corresponding target genes have been identified in the regulation of chemoresistance. For example, the interplays of long noncoding RNA HOXA11-AS1/miR-454/STAT3 drove chemoresistance of lung cancer [5]. miR-202 promoted cisplatin response in a RAS/MEK1 dependent manner [6]. Circulating miR-425 activated AKT and led to cisplatin resistance [7]. Due to one miRNA targets different mRNAs, while various miRNAs can regulate single mRNA, more knowledge is urgently needed to extend the knowledge of cisplatin resistance.
CTLA‐4, PD‐1, and PD‐L1 have been well studied and generated significant clinical benefits for advanced-stage cancer. PD-1 dominantly expresses in activated T cells and prevents T cells activation by interacting with PD-L1. PD-L1, the ligand of PD-1, expresses universally and increases in malignant cells [8]. Recent studies indicated that both PD-1 and PD-L1 contribute to cisplatin resistance. Kurimoto R and colleagues demonstrated that TGF-β/FGF2 promoted PD-L1 expression and conferred resistance against cisplatin [9]. Zhang P et al. found that PD-L1 reduction attenuated cisplatin resistance in lung cancer cells [10]. Accumulating data suggest that multiple signaling pathways merge in PD-L1 and confer lung cancer resistance against cisplatin afterward. Therefore, investigation of PD-L1-mediated cisplatin resistance is expected to provide a novel rationale for treating refractory lung cancer.
In the present study, we aimed to figure out the association of miR-526b-3p, STAT3, and PD-L1 in cisplatin-resistant lung cancer by collecting clinical information. We further attempted to clarify the details of miR-526b-3p/STAT3/PD-L1 signaling pathways-mediated resistance by conducting gain-of-function and loss-of-function experiments.
Material and methods
Patients’ recruitment and tissue samples collection
One hundred non-small-cell lung cancer patients who had received chemotherapy were recruited into the present study. Those patients who were diagnosed with synchronous distant metastasis, received targeted therapy, or immune checkpoint inhibitors were excluded. The cancer tissues were reviewed by two independent pathologists, and the complete pathological information was collected (Table 1). Written consent for research and publication was obtained from each participant. The study was approved by the Medical Ethics Committee of Shengjing Hospital of China Medical University.
miRNA microarray analysis
RNA gained from tumor tissues was used for custom miProfileTM Cancer miRNA qPCR Array (Cat. No. QM001-E, GeneCopoeia, Rockville, MD, USA). The methods for gathering RNA and synthesizing cDNA were described in the qRT-PCR section. qPCR arrays were conducted following the vendor’s protocol.
Cell lines and reagents
Human epithelial virus-transformed lung bronchus cell line BEAS-2B (ATCC@CRL-9609TM), human lung carcinoma cell LINE H1975 (ATCC@CRL-5908TM), A549 (ATCC@CRL-185TM) were provided by American Type Culture Collection (Manassas, VA, USA). PC-9 (formerly known as PC-14, Code: 90071810) was supplied by the European Collection of Cell Cultures (ECACC; Salisbury, United Kingdom). Cisplatin-resistant A549 (A549/DDP) and cisplatin-PC-9 (PC-9/DDP) were gifts from Dr. Xuejun Guo. BEAS-2B cells were grown in RPMI-1640 with 10% fetal bovine serum. The rest cells were cultured in Dulbecco’s modified Eagle medium (DMEM) containing 10% fetal bovine serum. Cisplatin-resistant cells were maintained with DMEM plus 0.1 μg/ml cisplatin as previously described [11]. Medium and serum were obtained from Sigma-Aldrich (St. Louis, MO, USA). Mycoplasma Detection Kit (Cat.No.MP0025, Sigma-Aldrich) was used to detect Mycoplasma during cell culture.
Cisplatin (Cat.No.C2210000) was purchased from Sigma-Aldrich. miR-526b-3p, miR-526b-3p inhibitor, and empty vector pEZX-MR04 were purchased from GeneCopoeia. pDONR223_STAT3_WT was a gift from Jesse Boehm & William Hahn & David Root (Addgene plasmid # 82235; http://n2t.net/addgene:82235; RRID: Addgene_82235) [12]. Vector pDONR223 was generated by Invitrogen (Cat.No.12536017, Carlsbad, CA, USA).
Putative miRNA and mRNA interactions
The candidate sponge mRNAs for miRNAs were predicted by miRDB online database (http://www.mirbase.org/) [13, 14].
Cell growth assay
MTT (3-[4,5- dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide) based-colorimetric assays were carried out adapted to the manufacturer’s protocol (Sigma-Aldrich, St. Louis, MO, USA). A total of 0.8 × 104 cells were plated in 96-well plates with 0.1 ml DMEM. Cells were treated with cisplatin at different doses. Finally, cells were incubated with ten μl MTT solution (5 mg/ml) for four hours at 37 ˚C prior to analysis. The optical densities were read at 490 nm using a Microplate Reader (Thermo Fisher Scientific, Waltham, MA, USA).
Migration assay
Transwell migration assay was performed using modified Boyden chambers (Sigma-Aldrich). A total of 4 × 104 cells in serum-free medium were plated in the upper room of each chamber, whereas the lower room was filled with medium supplemented with 10% fetal bovine serum. After incubating for 18 h, cells in the upper compartments were discarded. In contrast, the migrated cells in the lower parts were stained with 1% crystal violet solution, followed by counting under Olympus microscope CX23 (Shinjuku, Tokyo, Japan).
Western blots
Protein was collected by whole-cell lysis buffer. Forty micrograms protein was resolved with SDS-PAGE gel and was transferred to Polyvinylidene fluoride membranes (Sigma-Aldrich). The membranes were blocked with 5% skim milk for 1 h and incubated with the following primary antibodies overnight at a dilution of 1:1000. STAT3 (Cat.No.9139), PD-L1 (Cat.No.13684), c-Myc (Cat.No.5605), MDR1 (Cat.No.13342), and GAPDH (Cat.No.5174) were purchased from Cell Signaling Technology (Danvers, MA, USA). The membranes were washed with TBST and incubated with HRP-conjugated secondary antibodies (Cell Signaling Technology) at a dilution of 1:2000 for 1 h. Protein was visualized using the Western Lighting Ultra (Thermo Fisher Scientific).
Real-time quantitative polymerase chain reaction (RT-qPCR)
RNA was extracted by RNA isolation kit (Cat.No.83913, Sigma-Aldrich) per the manufacturer’s protocol. Two hundred fifty nanograms amount of RNA was reverse transcribed to cDNA by a Reverse Transcription Reaction kit (Qiagen, Hilden, Germany). RT-qPCR was carried out using the LightCycler®480 System (Roche, Basel, Switzerland). PCR was conducted as follows: 40 cycles of 94 °C for 18 s, 60 °C for 15 s, and 72 °C for 30 s. All procedures were repeated independently. Gene expression was normalized to GAPDH using the 2−ΔΔCq method [15]. All primers were synthesized by Genechem (Shanghai, China). The primer sequences for detecting the indicated genes were listed below.
miR-526b-3p, F,5′-CTCTTGAGGGAAGCACT-3′; R, 5′-GAACATGTCTGCGTATCTC-3′. STAT3, F,5′-CTTTGAGACCGAGGTGTATCACC-3′; R, 5′-GGTCAGCATGTTGTACCACAGG-3′. PD-L1, forward,5′-TGCCGACTACAAGCGAATTACTG-3′; R, 5′-CTGCTTGTCCAGATGACTTCGG-3′. MDR1, forward,5′-GCTGTCAAGGAAGCCAATGCCT-3′; R, 5′-TGCAATGGCGATCCTCTGCTTC-3′. c-Myc, forward,5′-CCTGGTGCTCCATGAGGAGAC-3′; R, 5′-CAGACTCTGACCTTTTGCCAGG-3′. GAPDH, F,5′-GTCTCCTCTGACTTCAACAGCG-3′; R, 5′-ACCACCCTGTTGCTGTAGCCAA-3′.
Luciferase reporter assay
1-1020 of 3′untranslation region of STAT3 was inserted into pDONR223. miR-526b-3p or miR-526b-3p mutant was inserted into pmirGLO (Cat.No. E1330, Promega, Madison, WI, USA) according to the vendor’s instructions. Twenty-four hours before transfection, cells were plated in 96-well dish plates at a density of 2 × 104 per well. pmirGLO-miR-526b-3p, pmirGLO-miR-526b-3p mutant (miR-526b-3pMUT) along with pDONR223_STAT3 3’UTR were introduced into cells prior to dual-luciferase reporter assay. Luciferase activity was determined post-24 h transfection with the Luc-Pair™ Duo-Luciferase Assay Kit 2.0 (GeneCopoeia).
Immunohistochemical staining
The STAT3 primary antibody (Cat.No.9139) was used to detect STAT3 expression in formalin-fixed, paraffin-embedded tissues according to the previous protocol [16]. Briefly, antigen was retrieved in citrate buffer (10 mM, pH 6.0), washed with phosphate-buffered saline, and exposed to 3% hydrogen peroxide. Slides were incubated with anti-STAT3 at a dilution of 1:200 at room temperature for one hour. ElivisionTM plus Polymer HRP (Mouse/Rabbit) IHC Kit (Cat.No.9901, Maxin, Fuzhou, China) was used to visualize STAT3 stains adapted to the vendor’s instruction.
A549 metastatic xenograft models
Five-week-old BALB/c male nude mice were purchased from Beijing Laboratory Animal Research Center (Beijing, China). Mice were housed in conditions under the guidelines of the experimental animal center of Shengjing Hospital. A total of 5 × 105 A549 cells were intravenously injected into fifteen nude mice. Mice were separated into three groups randomly (five per group) seven days later. Twenty-eight days post-injection, mice were sacrificed with carbon dioxide inhalation. Hematoxylin and eosin staining was conducted according to the standard procedure afterward [17]. Additionally, gene expression was accessed by Western blots and qPCR assay. All studies were approved by the medical ethics committee of Shengjing hospital of China Medical University. We complied with the guideline of the Experimental Animal Center of Shengjing Hospital of China Medical University during the generation of animal models.
Statistical analysis
Data in graphs were shown as mean ± standard deviation (SD). The association of miR-526b-3p and STAT3 was analyzed by Spearman’s correlation coefficient test. The differences among groups were evaluated by one-way or two-way Analysis of Variance, following by a Sidak’s multiple comparisons test. P < 0.05 indicated statistical significance. Statistical analysis was performed by using GraphPad version 8.1 (San Diego, CA, USA).
Results
miR-526b-3p decreases in cisplatin-resistant lung cancer
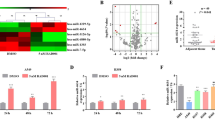
To explore the differential expression of miRNAs in cisplatin-resistant lung cancer, we conducted miRNA arrays with three cisplatin-resistant and three cisplatin-sensitive specimens. Fig 1a exhibited that miR-526b-3p dropped in cisplatin-resistant tissues. We further demonstrated that miR-526b-3p fell significantly in 50 cisplatin-resistant cases compared to that in cisplatin-sensitive cases (Fig. 1b). The results in Fig. 1c showed that miR-526b-3p declined in lung cancer cell lines compared to the immortalized human non-tumorigenic lung epithelial cell line BEAS-2B. We selected A549 and PC-9 as cell models in the following in-vitro experiments due to there was no significant difference in miR-526b-3p expression.
a The differential expression of miRNAs between cisplatin-sensitive and cisplatin-resistant lung cancer is analyzed by miRNA microarray. The color indicates the fold change of the target gene normalized to the corresponding housekeeping genes. b, c miR-526b-3p expression in tumor specimens and cell lines is determined by qRT-PCR. adj adjacent tissues; sen cisplatin-sensitive tumor tissues; res cisplatin-resistance tumor tissues. *, P < 0.05 vs. adjacent tissues or BEAS-2B cells. #, P < 0.05 vs. cisplatin-sensitive tissues.
miR-526b-3p suppresses lung cancer malignancy
To investigate the effects of miR-526b-3p in lung cancer cells, we introduced miR-526b-3p mimic (miR-526) and miR-526b-3p inhibitor (miR-526inh) into A549 and PC-9 cells. The results in Fig. 2a exhibited that expression of miR-526b-3p increased in cells post-transfection of miR-526b-3p mimic, while that decreased in cells with miR-526b-3p inhibitor. Besides, we introduced miR-526b-3p mimic into cisplatin-resistant cells, following by exposure to cisplatin. Fig 2b demonstrated that the IC50 values against cisplatin in the miR-526b-3p group were reduced compared to the vector. Besides, ectopic expression of miR-526b-3p suppressed cell viability while miR-526b-3p inhibitor promoted cell viability (Fig. 2c). Moreover, miR-526b-3p overexpression repressed cell migration, whereas miR-526b-3p knockdown enhanced cell migration (Fig. 2d). The results demonstrated that miR-526b-3p was a tumor suppressor in cisplatin-resistant lung cancer.
a The expression of miR-526b-3pin cells is accessed by qRT-PCR. b The viability of cells with miR-526b-3p post cisplatin exposure is determined by MTT assay. c The growth of the indicated cells is detected by MTT assay. d The migration of the indicated cells is determined by a transwell assay. *, P < 0.05 vs. control.
STAT3 is one of the targets of miR-526b-3p and increases in cisplatin-resistant lung cancer
We used miRDB database to predict potential candidates of miR-526b-3p. Signal Transducer and Activator of Transcription 3 (STAT3) exhibited the most upregulation among hundreds of candidate genes (Supplementary Figure). We, therefore, selected STAT3 to investigate further. Fig 3a showed the schematic diagram of the binding sites between miR-526b-3p and STAT3 3′UTR. We generated the miR-526b-3p (miR-526b-3pmut) mutant, which was insufficient to adhere to the 3′UTR of STAT3. The 3′UTR of STAT3 plus miR-526b-3p, or miR-526b-3pmut, were introduced into cells, followed by dual-luciferase assays. The results in Fig. 3b showed that the 3′UTR of STAT3 changed little in the miR-526b-3pmut group, while that in the miR-526b-3p group declined compared to control. In addition, we detected the expression of STAT3 in lung cancer. Fig 3c exhibited that the STAT3 expression in cisplatin-resistant lung cancer was higher than that in cisplatin-sensitive specimens. We also examined STAT3 expression in cancer tissues by qRT-PCR. The results were in line with those of immunohistochemical staining (Fig. 3d). Furthermore, STAT3 expression was negatively correlated with miR-526b-3p (Fig. 3e). The results indicated that miR-526b-3p inhibited STAT3 in cisplatin-resistant lung cancer.
a The putative binding sites of STAT3 3′UTR and miR-526b-3p are shown. b The relative dual-luciferase activity of STAT3 3′UTR in the indicated cells is measured. c The representative images of STAT3 expression in lung cancer tissues are shown. Sensitive, cisplatin sensitive. Resistant, cisplatin-resistant. Scale bar, 50 µm. d The expression of STAT3 in lung cancer tissues is accessed by qRT-PCR. *, P < 0.05 vs. adjacent tissues. #, P < 0.05 vs. cisplatin-sensitive tissues. e The correlation of STAT3 and miR-526b-3p is estimated by Spearman’s correlation coefficient. *, P < 0.05.
miR-526b-3p/STAT3 axis regulated lung cancer malignancy
To explore the role of the miR-526b-3p/STAT3 axis in the regulation of lung cancer malignancy, we introduced miR-526b-3p and STAT3 into cells. Fig 4a showed that STAT3 expression fell in cells post miR-526b-3p transfection, whereas that augmented in cells with the combination. The relative fluorescent unit of rhodamine123 in the miR-526b-3p group was reduced, whereas that in the combo escalated compared to control (Fig. 4b), suggesting that the ectopic expression of STAT3 reversed the inhibition of miR-526b-3p in drug efflux. The viability of cells with miR-526b-3p declined, while that of cells with the combo increased (Fig. 4c). The results proved that STAT3overexpression attenuated the suppression of miR-526b-3p in cell growth. Besides, miR-526b-3p inhibited the expression of programmed death-ligand 1 (PD-L1), while the combo promoted PD-L1 expression (Fig. 4d, e). Because PD-1/PD-L1 interaction enhances the immune evasion in tumors by inhibiting CD8+ T cells [18], we wondered whether the population of CD8+ T cells was interfered with by the introduction of miR-526b-3p. The results in Fig. 4f exhibited that miR-526b-3p triggered the population of CD8+ T cells, whereas STAT3 interrupted the promotion. The motility of cells in the miR-526b-3p group was reduced while that in the combo amplified compared to control (Fig. 4g).
a miR-526b-3p and STAT3 expression is accessed by qRT-PCR. b The activity of MDR1 is measured by rhodamine123 efflux assay. RLU relative fluorescence of unit. c The viability of the indicated cells is accessed by MTT assay. d The expression of the indicated genes is detected by qRT-PCR and Western blot, separately. e Representative immunofluorescent images of STAT3 and PD-L1 in A549 cells are shown. f The percentage of CD8 positive T-cells in the indicated cells are accessed by flow cytometry analysis. g The motility of the indicated cells was detected by transwell migration assay. Scale bar, 100 µm. *, P < 0.05 vs control. #, P < 0.05 vs miR-526b-3p plus vector.
miR-526b-3p/STAT3 axis regulated cisplatin resistance in a PD-L1-dependent manner
Previous studies have shown that STAT3 promotes PD-L1 expression, contributing to chemoresistance of breast cancer, head, and neck squamous cell carcinoma, and non-small-cell lung cancer [19,20,21]. We, therefore, investigated the effects of miR-526b-3p and STAT3 in PD-L1 expression. The results in Fig. 5a showed that the knockdown of miR-526b-3p promoted STAT3 and PD-L1 expression. Avelumab, a monoclonal antibody that targets PD-L1, significantly suppressed PD-L1 while having little effect in STAT3 or miR-526b-3p expression. The relative fluorescent unit of rhodamine123 in the miR-526b-3p inhibitor group increased, whereas that in the avelumab group dropped compared to control. Notably, avelumab repressed the effects of miR-526b-3p inhibitor in drug efflux (Fig. 5b). In addition, the population of CD8+ T cells in the miR-526b-3p inhibitor group fell, whereas that in the combination was similar to the avelumab group (Fig. 5c). Further, the viability of miR-526b-3p-knockdown cells developed while that of the combination group remained the same as the avelumab group (Fig. 5d). Moreover, miR-526b-3p inhibitor enhanced cell migration while avelumab repressed cell motility. Importantly, the cell migration of the combo group was identical to that of avelumab (Fig. 5e). The results indicated that avelumab treatment abolished miR-526b-3p-mediated anti-tumor effects. Thus, PD-L1 was a new downstream target of miR-526b-3p/STAT3 mediated cisplatin of lung cancer.
a Expression of miR-526b-3p, STAT3, and PD-L1 is accessed by qRT-PCR and western blot, separately. b The activity of MDR1 is measured by rhodamine123 efflux assay. RLU relative fluorescence of unit. c The percentage of CD8 positive T-cells in the indicated cells are accessed by flow cytometry analysis. d The viability of the indicated cells is accessed by MTT assay. e The motility of the indicated cells is detected by transwell migration assay. Scale bar, 100 µm. *, P < 0.05 vs. control. ns no significance vs avelumab.
miR-526b-3p/STAT3 regulated cancer metastasis in vivo
We generated A549 mouse metastatic models with cells carrying control, miR-526b-3p, miR-526b-3p plus STAT3. Lung metastasis was validated by hematoxylin-eosin stains (Fig. 6a). The results in Fig. 6b demonstrated that PD-L1 expression declined in the miR-526b-3p group, while that increased in the combination group. Besides, we found the oncogene c-Myc and the multidrug resistance regulator MDR1 decreased in the miR-526b-3p group, whereas those increased in the combo group (Fig. 6c). Thus, the observations obtained in the animal models were in line with the results gained in vitro.
a The representative images of lung metastasis by hematoxylin and eosin stains are shown. Scale bar, 20 µm. b The representative images of PD-L1 expression by immunohistochemical stain are shown. Scale bar, 50 µm. c The expression of the indicated genes is determined by western blot and qRT-PCR, respectively. *, P < 0.05 vs. control. #, P < 0.05 vs miR-526b-3p plus vector.
Discussion
Chemotherapy remains the preferred and primary treatment for advanced lung cancer. Cisplatin is widely used as the first-line medicine. Unfortunately, the frequent development of resistance prevents the continuous use of cisplatin and leads to treatment failure. Multiple mechanisms, such as gene mutation, abnormal gene expression, and miRNA dysregulation, play vital roles in developing cisplatin resistance. Increasing data have shown that miRNA expression in cisplatin-resistant cancer cells and the corresponding parental cancer cells are distinct. We found that miR-526b-3p overexpression enhanced the response to cisplatin while attenuating cell growth and migration in cisplatin-resistant lung cancer cells. The observations that miR-526b-3p as a tumor suppressor were supported by previous studies. For example, Ming W et al. demonstrated that miR-526b-3p targeted WEE1, leading to glioma regression [22]. Furthermore, Zhang R et al. proved that miR-526b-3p retarded colon cancer metastasis by suppressing HIF1α [23]. Besides, the ectopic expression of miR-526b-3p interrupted malignancies by forming complexes with long noncoding RNAs and mRNAs [24,25,26].
The aberrant activation of STAT3 is correlated to poor clinical outcomes of lung cancer [27]. The dysregulation of fibroblast growth factor receptor, vascular endothelial growth factor, IL-6, and extracellular signal-regulated kinase contribute to hyperactivation of STAT3. Particularly, STAT3 is one of the pivotal factors that cause cisplatin resistance. Tremendous studies have indicated that STAT3 is a promising therapeutic target for conquering cisplatin resistance [28, 29]. Recently, various miRNAs are emerging as critical regulators of STAT3. For example, miR-608 attenuated lung cancer malignancy in a JANK2/STAT3 dependent manner [30]. miR-146b-3p promoted cervical cancer cell proliferation and migration via STAT3 and AKT signaling pathways [31]. We demonstrated that miR-526b-3p directly targeted STAT3 and subsequently inhibited the expression of PD-L1, c-Myc, and MDR1. Apart from miR-526b-3p, STAT3 expression is regulated by different miRNA. Previous research has shown that miR-296-5p, miR-106-5p, miR-10b, and miR-21 target STAT3 by adhering to STAT3 3’UTR, contributing to cisplatin resistance [32,33,34,35]. The reasons for the divergence of miRNA-STAT3 interactions are not fully understood yet. The alternative genome in various cancer can be one of the explanations.
Previously, Shen M et al. proved that Ataxia Telangiectasia Mutated (ATM) promoted PD-L1 expression by enhancing JAK/STAT3 signaling cascades in lung cancer [36]. Zhang P et al. revealed that the IL-6/STAT3 axis conferred head and neck squamous cell carcinoma resistance against cisplatin [37]. Furthermore, Lei Z et al. found that the combination of anti-PD-L1 Atezolizumab and anti-vascular endothelial growth factor Bevacizumab prevented cisplatin-resistant ovarian cancer progression by suppressing STAT3-induced epithelial-mesenchymal transition [38]. The results mentioned above were consistent with our present results, indicating that STAT3/PD-L1-mediated cisplatin resistance is universal. PD-L1 elevates in various cancer cells post-chemotherapy and subsequently promotes cancer progression by facilitating immune evasion and chemoresistance [39]. A previous study showed that let-7-inhibited PD-L1 expression reactivated CD8+ T cells and reversed cisplatin resistance of non-small-cell lung cancer [40]. We demonstrated that the avelumab treatment abrogated the effects of miR-526b-3p inhibitor, providing new clues to draw a global picture of PD-L1-mediated chemoresistance.
Briefly, miR-526b-3p targeted STAT3 and inhibited cell growth, migration, and cisplatin resistance in a PD-L1-dependent manner. miR-526b-3p may be a potential therapeutic target for improving the anti-tumor effects of chemo-immunotherapy combination.
Data availability
The RT-PCR profiling analysis of the arrays is curated by Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE168707).
References
Ghosh S. Cisplatin: the first metal based anticancer drug. Bioorg Chem. 2019;88:102925.
Sun CY, Nie J, Huang JP, Zheng GJ, Feng B. Targeting STAT3 inhibition to reverse cisplatin resistance. Biomed Pharmacother. 2019;117:109135.
Jin L, Chun J, Pan C, Li D, Lin R, Alesi GN, et al. MAST1 drives cisplatin resistance in human cancers by rewiring cRaf-independent MEK activation. Cancer Cell. 2018;34:315–30e7.
Liang F, Ren C, Wang J, Wang S, Yang L, Han X, et al. The crosstalk between STAT3 and p53/RAS signaling controls cancer cell metastasis and cisplatin resistance via the Slug/MAPK/PI3K/AKT-mediated regulation of EMT and autophagy. Oncogenesis. 2019;8:59.
Zhao X, Li X, Zhou L, Ni J, Yan W, Ma R. et al. LncRNA HOXA11-AS drives cisplatin resistance of human LUAD cells via modulating miR-454-3p/Stat3. Cancer Sci.2018;109:3068–79.
Sun W, Ping W, Tian Y, Zou W, Liu J, Zu Y. miR-202 enhances the anti-tumor effect of cisplatin on non-small cell lung cancer by targeting the Ras/MAPK pathway. Cell Physiol Biochem. 2018;51:2160–71.
Ma Y, Yuwen D, Chen J, Zheng B, Gao J, Fan M, et al. Exosomal transfer of cisplatin-induced miR-425-3p confers cisplatin resistance in NSCLC through activating autophagy. Int J Nanomed. 2019;14:8121–32.
Xia L, Liu Y, Wang Y. PD-1/PD-L1 blockade therapy in advanced non-small-cell lung cancer: current status and future directions. Oncologist. 2019;24 Suppl 1:S31–S41.
Kurimoto R, Iwasawa S, Ebata T, Ishiwata T, Sekine I, Tada Y, et al. Drug resistance originating from a TGF-beta/FGF-2-driven epithelial-to-mesenchymal transition and its reversion in human lung adenocarcinoma cell lines harboring an EGFR mutation. Int J Oncol. 2016;48:1825–36.
Zhang P, Ma Y, Lv C, Huang M, Li M, Dong B, et al. Upregulation of programmed cell death ligand 1 promotes resistance response in non-small-cell lung cancer patients treated with neo-adjuvant chemotherapy. Cancer Sci. 2016;107:1563–71.
Song L, Cui Z, Guo X. Comprehensive analysis of circular RNA expression profiles in cisplatin-resistant non-small cell lung cancer cell lines. Acta Biochim Biophys Sin. 2020;52:944–53.
Kim E, Ilic N, Shrestha Y, Zou L, Kamburov A, Zhu C, et al. Systematic functional interrogation of rare cancer variants identifies oncogenic alleles. Cancer Discov. 2016;6:714–26.
Liu W, Wang X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 2019;20:18.
Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43:D146–52.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8.
Lee SW, Ahn YY, Kim YS, Kang SB, Nam SW, Lee DS, et al. The immunohistochemical expression of STAT3, Bcl-xL, and MMP-2 proteins in colon adenoma and adenocarcinoma. Gut Liver. 2012;6:45–51.
Fischer AH, Jacobson KA, Rose J, Zeller R. Hematoxylin and eosin staining of tissue and cell sections. CSH Protoc. 2008;2008:pdb prot4986.
Hobo W, Maas F, Adisty N, de Witte T, Schaap N, van der Voort R, et al. siRNA silencing of PD-L1 and PD-L2 on dendritic cells augments expansion and function of minor histocompatibility antigen-specific CD8+ T cells. Blood. 2010;116:4501–11.
Gao W, Wen H, Liang L, Dong X, Du R, Zhou W, et al. IL20RA signaling enhances stemness and promotes the formation of an immunosuppressive microenvironment in breast cancer. Theranostics. 2021;11:2564–80.
Lee Y, Shin JH, Longmire M, Wang H, Kohrt HE, Chang HY. et al.CD44+ cells in head and neck squamous cell carcinoma suppress T-cell-mediated immunity by selective constitutive and inducible expression of PD-L1.Clin Cancer Res. 2016;22:3571–81.
Tang D, Zhao D, Wu Y, Yao R, Zhou L, Lu L. et al. The miR-3127-5p/p-STAT3 axis up-regulates PD-L1 inducing chemoresistance in non-small-cell lung cancer. J Cell Mol Med. 2018;22:3847–3856.
Wu M, Li X, Liu Q, Xie Y, Yuan J, Wanggou S. miR-526b-3p serves as a prognostic factor and regulates the proliferation, invasion, and migration of glioma through targeting WEE1. Cancer Manag Res. 2019;11:3099–110.
Zhang R, Zhao J, Xu J, Wang J, Jia J. miR-526b-3p functions as a tumor suppressor in colon cancer by regulating HIF-1alpha. Am J Transl Res. 2016;8:2783–9.
Yin G, Tian P, BuHe A, Yan W, Li T, Sun Z. LncRNA LINC00689 promotes the progression of gastric cancer through upregulation of ADAM9 by sponging miR-526b-3p. Cancer Manag Res. 2020;12:4227–39.
Yan F, Ma Y, Liu L, Li L, Deng J, Sun J. Long noncoding RNA HOXD-AS1 promotes the proliferation, migration, and invasion of colorectal cancer via the miR-526b-3p/CCND1 axis. J Surg Res. 2020;255:525–35.
Zhan WL, Gao N, Tu GL, Tang H, Gao L, Xia Y. LncRNA LINC00689 promotes the tumorigenesis of glioma via mediation of miR-526b-3p/IGF2BP1 axis. Neuromolecular Med. 2021. https://doi.org/10.1007/s12017-020-08635-x.
Tong M, Wang J, Jiang N, Pan H, Li D. Correlation between p-STAT3 overexpression and prognosis in lung cancer: a systematic review and meta-analysis. PLoS One. 2017;12:e0182282.
Lee CH, Yang JR, Chen CY, Tsai MH, Hung PF, Chen SJ. et al. Novel STAT3 inhibitor LDOC1 targets phospho-JAK2 for degradation by interacting with LNX1 and regulates the aggressiveness of lung cancer. Cancers. 2019;11:63
Guo S, Luo W, Liu L, Pang X, Zhu H, Liu A, et al. Isocryptotanshinone, a STAT3 inhibitor, induces apoptosis and pro-death autophagy in A549 lung cancer cells. J Drug Target. 2016;24:934–42.
Xu W, Sun D, Wang Y, Zheng X, Li Y, Xia Y, et al. Inhibitory effect of microRNA-608 on lung cancer cell proliferation, migration, and invasion by targeting BRD4 through the JAK2/STAT3 pathway. Bosn J Basic Med Sci. 2020;20:347–56.
Yao S, Xu J, Zhao K, Song P, Yan Q, Fan W, et al. Down-regulation of HPGD by miR-146b-3p promotes cervical cancer cell proliferation, migration, and anchorage-independent growth through activation of STAT3 and AKT pathways. Cell Death Dis. 2018;9:1055.
Dong Y, Xu T, Zhong S, Wang B, Zhang H, Wang X, et al. Circ_0076305 regulates cisplatin resistance of non-small cell lung cancer via positively modulating STAT3 by sponging miR-296-5p. Life Sci. 2019;239:116984.
Guo J, Dou D, Zhang T, Wang B. HOTAIR promotes cisplatin resistance of osteosarcoma cells by regulating cell proliferation, invasion, and apoptosis via miR-106a-5p/STAT3 axis. Cell Transpl. 2020;29:963689720948447.
Tan WX, Sun G, Shangguan MY, Gui Z, Bao Y, Li YF, et al. Novel role of lncRNA CHRF in cisplatin resistance of ovarian cancer is mediated by miR-10b induced EMT and STAT3 signaling. Sci Rep. 2020;10:14768.
Yao T, Lu R, Zhang J, Fang X, Fan L, Huang C, et al. Growth arrest-specific 5 attenuates cisplatin-induced apoptosis in cervical cancer by regulating STAT3 signaling via miR-21. J Cell Physiol. 2019;234:9605–15.
Shen M, Xu Z, Xu W, Jiang K, Zhang F, Ding Q, et al. Inhibition of ATM reverses EMT and decreases metastatic potential of cisplatin-resistant lung cancer cells through JAK/STAT3/PD-L1 pathway. J Exp Clin Cancer Res. 2019;38:149.
Zhang P, Liu J, Li W, Li S, Han X. Lactoferricin B reverses cisplatin resistance in head and neck squamous cell carcinoma cells through targeting PD-L1. Cancer Med. 2018;7:3178–3187.
Zhang L, Chen Y, Li F, Bao L, Liu W. Atezolizumab and bevacizumab attenuate cisplatin resistant ovarian cancer cells progression synergistically via suppressing epithelial-mesenchymal transition. Front Immunol. 2019;10:867
de Mel S, Hue SS, Jeyasekharan AD, Chng WJ, Ng SB. Molecular pathogenic pathways in extranodal NK/T cell lymphoma. J Hematol Oncol. 2019;12:33.
Hong W, Xue M, Jiang J, Zhang Y, Gao X. Circular RNA circ-CPA4/ let-7 miRNA/PD-L1 axis regulates cell growth, stemness, drug resistance, and immune evasion in non-small cell lung cancer (NSCLC). J Exp Clin Cancer Res. 2020;39:149.
Acknowledgements
The PC-9 cell line was supplied by the European Collection of Cell Cultures (ECACC; Salisbury, United Kingdom) as catalog number 90071810 and was purchased from CellBank Australia (Westmead, NSW, Australia). Cisplatin-resistant A549 (A549/DDP) and cisplatin-PC-9 (PC-9/DDP) were gifts from Dr. Xuejun Guo.
Funding
The author(s) received no specific funding for this work.
Author information
Authors and Affiliations
Contributions
KC and WY performed study concept and design; KC, YW, and AL performed development of methodology, writing, and revision of the paper; YX, and AL provided acquisition, analysis, and interpretation of data, and statistical analysis. All authors read and approved the final paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics
Written consent for research and publication was obtained from each participant. The study was approved by the Medical Ethics Committee of Shengjing Hospital of China Medical University (20180164335-5). All procedures in the study were performed in accordance with the Declaration of Helsinki.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by R. Mantovani
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chen, Kb., Yang, W., Xuan, Y. et al. miR-526b-3p inhibits lung cancer cisplatin-resistance and metastasis by inhibiting STAT3-promoted PD-L1. Cell Death Dis 12, 748 (2021). https://doi.org/10.1038/s41419-021-04033-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-021-04033-8
This article is cited by
-
MicroRNAs as regulators of immune checkpoints in cancer immunotherapy: targeting PD-1/PD-L1 and CTLA-4 pathways
Cancer Cell International (2024)
-
miR-185-5p May Modulate the Chemosensitivity of LUSC to Cisplatin via Targeting PCDHA11: Multi-omics Analysis and Experimental Validation
Biochemical Genetics (2024)