Abstract
Karonudib (TH1579) is a novel compound that exerts anti-tumor activities and has recently entered phase I clinical testing. The aim of this study was to conduct a pre-clinical trial in patient-derived xenografts to identify the possible biomarkers of response or resistance that could guide inclusion of patients suffering from metastatic melanoma in phase II clinical trials. Patient-derived xenografts from 31 melanoma patients with metastatic disease were treated with karonudib or a vehicle for 18 days. Treatment responses were followed by measuring tumor sizes, and the models were categorized in the response groups. Tumors were harvested and processed for RNA sequencing and protein analysis. To investigate the effect of karonudib on T-cell-mediated anti-tumor activities, tumor-infiltrating T cells were injected in mice carrying autologous tumors and the mice treated with karonudib. We show that karonudib has heterogeneous anti-tumor effect on metastatic melanoma. Thus, based on the treatment responses, we could divide the 31 patient-derived xenografts in three treatment groups: progression group (32%), suppression group (42%), and regression group (26%). Furthermore, we show that karonudib has anti-tumor effect, irrespective of major melanoma driver mutations. Also, we identify high expression of ABCB1, which codes for p-gp pumps as a resistance biomarker. Finally, we show that karonudib treatment does not hamper T-cell-mediated anti-tumor responses. These findings can be used to guide future use of karonudib in clinical use with a potential approach as precision medicine.
Similar content being viewed by others

Introduction
Cutaneous melanoma is the most aggressive form of skin cancer and is often fatal in metastatic stages1. Recent advances in melanoma genetics and how melanoma cells escape immune attack have resulted in the development of targeted therapies inhibiting the MAPK pathway as well as immunotherapies. These treatments have improved the overall survival and sometimes have resulted in patients possibly being cured from their disease2. Despite these successes, new treatments are needed, since the majority of patients are still not cured.
The recently developed inhibitor, karonudib (TH1579), was designed to inhibit the oxidized nucleotide-sanitizing enzyme, MTH1 (encoded by NUDT1)3, 4. The compound has shown promising anti-tumor effect both in vitro and in vivo4 and has initiated phase I clinical testing in cancer patients with advanced solid malignancies (NCT03036228). Lately, known anti-cancer effects, such as microtubule inhibition, have been proposed for compounds containing the same scaffold as karonudib (TH588 and TH287) (unpublished observations)5,6,7. The exact mechanism of action of karonudib and the possible role of MTH1 in cancer is therefore under investigation.
Regardless of the mechanism of action of the compound, karonudib exhibits good cytotoxic effect in cancer cells both in vitro and in vivo, while being well tolerated in non-transformed cells3, 4. However, the extent of inter-patient heterogeneity of the cytotoxic effect has not been determined. Furthermore, no predictive biomarkers of response have been identified. These could be used to identify which patients may benefit the most from this treatment and which patients may be spared. Lastly, given the recent successes of immunotherapies in melanoma, karonudib needs to be assessed for its potential impact on T-cell immunity. Here we made use of our melanoma PDX platform8 and a novel immune oncology PDX model9 to assess the heterogeneous responses observed when treating metastatic melanoma tumors in multiple patients with karonudib. We show that karonudib has anti-tumor effect in a majority of PDX models, but some PDX models remain unaffected, which in some cases can be attributed to known anti-cancer drug resistance mechanisms. Reassuringly though, we also demonstrate that karonudib treatment does not hamper cytotoxic T-cell anti-tumor activities.
Results
Heterogeneous response to karonudib treatment in melanoma PDXes
We have previously shown that TH588 and karonudib can inhibit the tumor growth of one of our PDX models3, 4. However, the heterogeneous responses of metastatic melanoma have not been modeled. To that end, we designed a pre-clinical PDX trial consisting of tumor samples from 31 metastatic melanoma patients representing models of most subtypes (Supplemental table 1). All samples had been serially transplanted in NOG mice as previously described8. Each sample was transplanted in three mice, and the two fastest growing xenografts were divided into two treatment groups, thus utilizing one mouse per patient per treatment group (1 × 1 × 1) as previously described10. The groups were treated with either 90 mg/kg karonudib or with the vehicle. All mice tolerated the treatment, as evidenced by their stable weights throughout the treatment (Supplemental figures S2-S4). After 3 weeks of treatment, the mice were sacrificed and tumors were harvested. A piece from each tumor was embedded in paraffin for immunohistochemical staining analysis, and a piece of each vehicle-treated tumor was RNA sequenced for mutation and expression analysis (Fig. 1a).
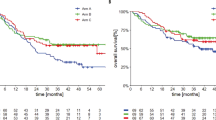
a Schematic overview of the experimental setup. Tumor biopsy from melanoma patient was serially transplanted twice in mice before being transplanted in mice treated with either karonudib or vehicle. For each patient, there was one mouse per treatment group, which was treated for 18 days before tumors were harvested and processed. Each tumor was snap-frozen for RNA analysis and embedded in paraffin for immunohistochemical straining. b Waterfall plot revealing the heterogenous response to karonudib. The samples are divided in three response groups, progression (blue), suppression (yellow), and regression (red). For treatment response catagorization, see Materials and methods. Comparison of average (±SEM) growth speed of karonudib treated vs. vehicle treated PDXes for (c) progression group, d suppression group, and e regression group. Individual growth curves are shown in supplemental figures S2-S4. f Kaplan–Meier graph showing the comparison of probability of progression-free survival in karonudib-treated mice (based on tumor doubling time) for the three response groups (see Materials and methods for criterion), Mantel–Cox test used for statistical analysis. g Quantification of immunohistochemical staining for the proliferation marker Ki-67 in vehicle vs. karonudib-treated PDXes, Student’s t test showing statistically significant difference between the two groups (p = 0.0476). h Images of the xenograft sections stained with Ki-67 and used for the quantification in (g)
After 3 weeks of treatment, heterogeneous responses to karonudib treatment were observed for the 31 PDX models (Fig. 1b and Supplemental figure S1-S4). Twenty-six percent (8/31) of the PDX models were regressed during karonudib treatment and thus, were categorized as the regression group. Forty-two percent (13/31) of the PDX models responded to karonudib treatment by reduced growth, compared with vehicle-treated tumors, and were categorized as the suppression group. Interestingly though, 32% (10/31) of the PDX models did not exhibit any reduced growth of karonudib-treated tumors, compared with vehicle; these were therefore categorized as the progression group.
To further investigate the treatment responses, the growth of each karonudib-treated PDX was compared with the growth of its matching vehicle-treated PDX (Fig. 1c–e). A significant difference in growth speed of the vehicle-treated PDXes was observed between the response groups, where the fastest growing PDXes were found in the suppression group. Furthermore, statistically significant difference was observed in the probability of progression-free survival (PFS), as based on tumor doubling time between the groups (p = 0.0039) (Fig. 1f). This markedly suppressed growth upon karonudib treatment correlated significantly (p = 0.03) to lower amount of Ki67 positive cells compared to the vehicle-treated mice in the suppression group, as analyzed with immunohistochemistry (Fig. 1g, h).
Karonudib has cytotoxic effect on melanoma tumors, irrespective of genotype
To investigate if the cytotoxic effect of karonudib was dependent on the genotype, the vehicle tumors were RNA sequenced and the mutation profile of the samples were analyzed. This analysis revealed BRAF and NRAS mutation in 65% (20/31) and 35% (11/31) of the samples, respectively. Comparison of the mutation profile between the response groups revealed that karonudib has cytotoxic effect in melanoma PDXes, irrespective of the presence of the genotype of the most common driver genes in melanoma, although a trend for an association between mutation in DDX3X and the regression group was observed (Fig. 2a and Supplemental figure S5A). However, the mutations in DDX3X were difficult to interpret, since these can be spread throughout the coding sequence and could possibly also arise by RNA editing.
Further investigation of the DDX3X mutation revealed increased PFS for karonudib-treated PDXes harboring the mutation when compared to the wild-type PDXes, even though not statistically significant (Supplemental figure S5b). Also, three different DDX3X missense mutations were revealed (Supplemental figure S5c and Supplemental table S2). To investigate if the impairment of the DDX3X protein had functional relevance when treating tumor cells with karonudib, the melanoma cell line MML-1 was transfected with a siRNA targeted against DDX3X and the cells treated with karonudib. A statistically significant lower expression of DDX3X was observed in the siDDX3X -transduced cells compared with the control-transduced cells (p = 0.0003; Supplemental figure S5d). DDX3X knockdown had a cytotoxic effect, and only 40% of the cells were alive 36 h after transfection (p < 0.0001; Supplemental figure S5e). When the DDX3X siRNA-transfected cells were treated with increased concentration of karonudib, a reduced number of live cells was observed when treated with 0.05 µM karonudib compared with the control (adjusted p value = 0.0002) (Supplemental figure S5f).
High expression of ABCB1 identified as a potential resistance biomarker
To screen for a predictive or a resistance biomarker for karonudib treatment, the association between the gene expression profile of the samples and response groups was investigated. Differential gene expression analysis was performed using the RNA-sequencing data from the vehicle-treated samples (for information regarding total reads per sample, see supplemental table S3). Comparison of the progression group to the suppression and the regression groups revealed differentially expressed genes, one of which was ABCB1 (Fig. 2b, c and supplemental table S4). Although highly expressed only in two of the samples in the progression group (p = 8.52*10−5 and q = 0.049 compared to the suppression group), it was of interest, since it encodes the multi-drug resistance (MDR) pump, also known as the p-glycoprotein pump (p-gp) (Fig. 3a).
a A waterfall plot showing treatment response for each of the PDX sample (see criterion in Materials and methods). Bars are color coded according to expression level of ABCB1 in the vehicle-treated PDXes, blue = 0–99 normalized reads, yellow = 100–1000 normalized reads, and red = >1000 normalized reads (for normalization method used see Materials and methods). b Quantitative analysis of ABCB1 expression in SK-MEL-2 cells after transduction with an ABCB1-expressing virus, compared with the control virus (±SD) (p < 0.0001), as analyzed with qRT-PCR. c Quantitative analysis of protein expression in the same cells, as b, analyzed with flow cytometry. d Quantitative analysis of activity of p-gp pumps using the p-gp substrate Rhodamine 123, as analyzed using flow cytometry, cells either treated with the p-gp inhibitor Elacridar (1 µM) or DMSO. e SK-MEL-2 cells transduced with ABCB1-expressing virus or control virus, treated with different concentrations of karonudib in combination with 0.1 µM Elacridar or DMSO for 48 h. Cells analyzed using flow cytometry, and the data shown as average of triplicate (±SD). Statistically significant rescue effect was observed when SK-MEL-2ABCB1 cells were treated with 0.5 µM karonudib (adjusted p value 2.6*10−8). f Immunohistochemical staining of ABCB1 in vehicle and karonudib-treated xenografts from the progression group
To address the functional importance of high ABCB1 expression, we transduced the melanoma cell line SK-MEL-2 with either an ABCB1-expressing virus or a control virus. Using quantitative real-time PCR, we observed statistically higher expression of ABCB1 in the ABCB1-transduced cells, compared to the control (p < 0.0001) (Fig. 3b). Furthermore, we verified that the high mRNA expression was translated to protein level, using flow cytometry analysis (Fig. 3c). The pumping activity of p-gp pumps can be followed by flow cytometry analysis of the p-gp substrate Rhodamine 123. Cells with functional pumps cultured in the presence of the substrate will pump out Rhodamine 123 and become negative for the stain over time. Our observation revealed high efficiency of the ABCB1-expressing melanoma cells to pump out Rhodamine 123. This activity could be inhibited with the p-gp pump inhibitor Elacridar (Fig. 3d). When treating the ABCB1-expressing cells with increasing concentration of karonudib, statistically significant less cytotoxic effect was observed for the ABCB1-expressing cells compared with the control cells (adjusted p value < 0.001). This protective effect could be reversed by using the p-gp pump inhibitor Elacridar (Fig. 3e). Finally, to assess if karonudib treatment affects ABCB1 expression or causes selection of ABCB1 high expressing cells, immunohistochemical staining of biopsies from four PDXes with high and one with low expression of ABCB1 mRNA (Fig. 3a) was performed. Only one PDX (M150330) showed somewhat higher expression of ABCB1 after karonudib treatment, suggesting that this is not a common mechanism of acquired resistance (Fig. 1f). Taken together, the data show that high expression of p-gp pumps can make cells less sensitive to karonudib treatment.
Karonudib treatment does not impair the T-cell-mediated anti-tumor response
Immunotherapy has recently proven to be an efficient treatment for patients suffering from metastatic cutaneous melanoma. New targeted therapies or chemotherapies therefore need to spare the effector cells, primarily cytotoxic T cells, if used in combination with immunotherapy. To inform on the potential usefulness of karonudib in the immune therapy era, we investigated if karonudib would affect the anti-tumor activity of cytotoxic T cells. First, we expanded tumor-infiltrating T lymphocytes (TILs) from patient biopsy M151002 and then added them to short-term cultures of tumor cells from the same patient in the presence or absence of karonudib. This resulted in degranulation of the T cells, as measured by increased CD107 positive cells (Fig. 4a) and increased secretion of IFN-gamma (Fig. 4b) when TILs were co-cultured with autologous tumor cells for 24 h. Reassuringly, no significant decrease was observed when the cells were cultured in the presence of karonudib. To test if this would cause anti-tumor effects in vivo, we used our recently developed method to develop immune humanized PDX models (PDXv2)9. PDX-generated M151002 tumor cells were injected in NOG mice or NOG mice, transgenic for human IL2 (hIL2-NOG). When tumor growth was confirmed, we injected TILs in the hIL2-NOG mice, and subsequently treated half of the NOG mice and half of the hIL2-NOG mice with karonudib. Again, karonudib treatment did not impair the anti-tumor activity of TILs. The response was assessed by both measuring the physical size of the tumors (Fig. 4c) and the bioluminescence from the luciferase-expressing tumor cells (Fig. 4d, e). Furthermore, we tested if karonudib impaired the anti-tumor effect of anti-CTLA-4 treatment in immunocompetent mice bearing mouse melanoma tumors. We transplanted B16F10 mouse melanoma cells subcutaneously in B6 mice and treated with vehicle, anti-CTLA-4, karonudib, or combination of CTLA-4 and karonudib. Tumors on mice treated with anti-CTLA-4 antibody alone showed good responses by suppression of tumor growth, which was not impaired by the combination treatment with karonudib (Fig. 4f). Taken together, this indicates that karonudib does not have obvious negative effect on T-cell-mediated anti-tumor effect.
a Quantitative analysis of the degranulation marker CD107 on TILs from patient sample M151002 grown with or without autologous tumor cells and with or without karonudib, data shown as average of triplicates (±SD). b Concentration of IFN-gamma in the media of the M15002 tumor cells cultured with or without autologous TILs and with or without karonudib. The ELISA data are shown as average of triplicates (±SD). c–e Size or luminescence signal of PDX tumors treated with vehicle (black), karonudib (red), injected with autologous TILs (blue) or karonudib + TILs (green), data are shown as average of four mice (±SEM). f B6 mice bearing tumors established from B16 mouse melanoma cell line, treated with either vehicle, CTLA4 antibody, karonudib, or combination of CTLA4 antibody and karonudib. Data in (f) shown as average tumor size of four mice (±SEM)
Conclusion
Here we show that the previously published anti-cancer effects of karonudib in one melanoma PDX4 can be demonstrated in a larger cohort of animal models. Importantly though, we also demonstrate that some models remain unaffected by karonudib, suggesting resistance mechanisms. The data therefore confirm the utility of PDX trials when preclinically assessing small molecule inhibitor efficacy10, 11. In the clinic, the response evaluation criteria in solid tumors (RECIST) criterion is used to evaluate the treatment response12. In a recent PDX trial paper and in a second paper re-analyzing the same data, a modified RECIST criterion is used to define the response groups10, 11. When using the RECIST criterion, the inherent growth speed of the tumor is not taken into account when assessing the treatment response. Since we used a matching vehicle-treated PDX for all karonudib-treated PDXes, it allowed us to take the inherent tumor growth speed into consideration when categorizing samples in the response groups (treated/control). We observed that the tumors with the fastest inherent growth speed also exhibited the biggest size difference between the treated and the untreated tumors. Using normal RECIST, these tumors would have been categorized as stable disease. Our study shows that such a stable disease would have clear clinical benefit in comparison to no treatment.
Interestingly, we observe that karonudib has anti-tumor effect on melanoma PDXes, irrespective of mutation status of major melanoma driver genes. Earlier work suggested that oncogenic KRAS-driven tumors would benefit from MTH1 expression13,14,15. We did not see any difference in sensitivity between BRAF- and NRAS-mutated tumors. This either means that the same rules do not apply in melanoma, that oncogenic KRAS and NRAS differ or that the mechanism of action of karonudib extends outside MTH1 inhibition, e.g., the proposed microtubule disruption caused by inhibitors of the same class as karonudib5,6,7.
We also observed that four PDX models in the cohort exhibited a mutation or variant in DDX3X. The fact that all these models were in the regression group was interesting. DDX3X is an RNA-binding protein and when mutated, it is known to impair global translation and induce stress granulation assembly16. Stress granules are normally formed in cells under acute stress, for example, oxidative stress, UV irradiation, and heat shock. Furthermore, it has been shown that cells treated with microtubule-targeting agents show difference in size and location of stress granules17. However, we cannot, at this time, state that DDX3X mutation is a predictive biomarker due to two reasons: First, the functional analysis hampered by DDX3X knockdown was lethal and therefore only additive to the effects of karonudib. Second, none of the observed DDX3X mutations have been described before, meaning that additional analyses are needed to draw a firm conclusion.
One of the aims of this study was to identify a predictive biomarker of response. High expression of p-glycoprotein (p-pg) is a well-known drug-resistance mechanism18. P-gp pumps are transmembrane proteins, which act as ATP-dependent drug-efflux pumps. They are known to recognize and excrete compounds, even compounds the cell has not been exposed to before19. Furthermore, it has been shown that MDR genes such as ABCB1 and ABCG2 are dispensable for mouse development20, suggesting that they might be suitable targets. Here, we observe that high-expressing ABCB1 cells are less sensitive to lower concentrations of karonudib than the control cells, which could be reversed by co-treating the cells with Elacridar (a known p-gp pump inhibitor18), establishing that karonudib is a p-gp substrate. That observation could be useful in the clinical setting for patients known to bear tumors with high p-gp pump expression, for example, in patients who have acquired resistance to chemotherapy21. Also, it raises the possibility of using p-gp pump inhibitors in combination with karonudib in the clinic. That combination treatment could be especially beneficial for melanoma patients, given the high expression of p-gp pumps in the blood–brain barrier, and that 10–20% of melanoma patients exhibit brain metastases.
It is likely that current treatment regiments only will cause durable responses or cures in subgroups of patients with melanoma. Current melanoma treatment includes both targeted therapy (BRAF and MEK inhibitors) as well as immune therapy (checkpoint block antibodies directed against PD1, PD-L1, and CTLA-4)2. Since durable responses to both these treatments have been correlated to abundance of tumor-infiltrating lymphocytes22, 23, it is imperative that novel treatments do not block the anti-tumor immunity. Hence, the fact that karonudib neither hinders adoptive T-cell transfer nor anti-CTLA-4 treatment in mice suggests that it can be used in combination with immune therapy.
Future use of karonudib is dependent on not only that it survives phase I clinical testing, but also that it causes clinically meaningful responses in patients. Most of the PDX models tested here reach a clinical response of stable disease. If this is reproduced in patients, combination treatments might be used to achieve tumor regression. Finding the right drug combinations (besides drug-pump inhibitors) is under active investigation.
Materials and methods
Ethical approvals
All experiments using patient material or research animals were performed according to the ethical approval provided by the Regional Human Ethics Board of Västra Götaland, Sweden #288-12 or by the Animal Ethics Board #2016-34, respectively.
Patient material
All samples were obtained from patients who had provided informed consent and were treated at the Department of Surgery, Sahlgrenska University Hospital, Gothenburg, Sweden.
Study design of the patient-derived xenograft clinical trial
Patient-derived xenografts were established from 31 metastatic melanoma patients, as previously described8. Each patient sample was transplanted in three mice. Palpable xenografts were measured three times weekly using a caliber and assigned to their treatment groups when they reached 50–100 mm3. The first mouse to reach the inclusion criterion was assigned to the karonudib treatment group, the second to the vehicle group, and the third mouse was excluded, utilizing the one mouse per patient per treatment group setup previously described10. Each mouse was treated for 18 days with either 90 mg/kg karonudib or the vehicle (20% w/v HPβCD in sodium acetate buffer pH 4.6) twice a day, three times per week. Treatment response was assessed by monitoring changes in tumor volume of the treated mouse and comparing to the tumor volume of the vehicle-treated mouse (treated/control (%)) using the following formulas:
\({\rm{tumor}}\,{\rm{volume}}\left( {{\rm{mm}}^3} \right)= \frac{{\rm{shorter}}\,{\rm{diameter}}^2\left( {\rm{mm}} \right) \times {\rm{longer}}\,{\rm{diameter}}{\rm{(mm)}}}{2}\)
and
\(T{\mathrm{/}}C\left( {\mathrm{\% }} \right) = \frac{{T_i - T_0}}{{C_i - C_0}}\) where Ti and Ci represents tumor sizes at the end of the treatment and T0 and C0 represents tumor sizes at treatment start. Response criterion was as follows; progression if T/C > 50%, suppression if T/C = 50–0%, and regression if T/C < 0%. Weight of mice and tumor size were measured three times per week. After 18 days, mice were sacrificed and the tumors harvested and weighted. Tumor pieces were processed and preserved as snap-frozen or embedded in paraffin for RNA or protein analysis, respectively.
RNA analysis
Tumors were dissociated using a Bullet Blender® and RNA was extracted using Nucleospin RNA kit (Machery-Nagel). Quantitative RNA analysis was conducted by synthesizing cDNA (iScript, BioRad) from the extracted RNA, which was quantified by qPCR (Kapa Biosystems). Primer sequences are available upon request. The extracted RNA was also used for RNA sequencing (SciLifeLabs NGS Core Facility, Stockholm, Sweden). Library was prepared using the Illumina TrueSeq with poly-A-selection and sequenced in a HiSeq 2500.
Immunohistochemistry
Fresh tumor pieces were fixed in 4% formalin, dehydrated, and embedded in paraffin. Next, 5-μm sections were made, mounted on glass slides, and dried over night at 37 °C. Rehydration and antigen retrieval were performed by pressure cooking in citrate buffer. Staining was performed with an auto-stainer (Autostainer Link 48, Dako). Primary antibody staining was done for 60 min at room temperature, secondary for 20 min, and horseradish peroxidase staining for 20 min. DAB (Diaminobenzidine) staining was used to stain the DNA and counterstaining was done using hematoxylin. Finally, the sections were dehydrated, mounted with Pertex, and imaged.
Antibodies
Anti-human Ki-67 antibody (DAKO) and anti-human ABCB1 antibody (Cell Signaling #13342).
Compounds
Karonudib (TH1579) was synthesized at the Karolinska Institute according to previously published synthesis schemes (WO2015187088A1). Elacridar (GF120918) was bought from Selleck Chem (selleckchem.com).
Bioinformatics
Alignment and pre-processing for variant calling
Raw reads were aligned to a combined human (hg19) and mouse (mm10) reference using STAR24 2.5.2b with default parameters. The index file for STAR was augmented with splice junctions from a concatenated human GENCODE25 version 17 and mouse GENCODE version M726 reference annotation. Reads mapping to human and mouse chromosomes, respectively, were then extracted using Samtools27 0.1.19. For reads mapping to both organisms, Samtools and the FilterSamReads module of Picard 1.109 (github.com/broadinstitute/picard) were then used to discard those that were not primary alignments with respect to the human reference. This strategy was motivated by the desire to minimize false positives in variant calling due to potential mouse contamination in the tumor samples.
Variant calling and filtering
Single-nucleotide variants and indels were called by first using the mpileup module of Samtools with disabled BAQ computation, a supplied list of regions for the genes of interest as well as the hg19 reference. The resulting output was then supplied to the VarScan28 2.3.9 tool mpileup2cns, for which the settings “min-coverage 2”, “min-reads2 2”, “min-var-freq 0.01”, and “min-avg-qual 15” were used. This very sensitive option was motivated by the desire to minimize the risk of false negatives owing to the lowly expressed genes and allelic imbalance, combined with the small list of genes being considered for calling. To further increase sensitivity, the two steps above were performed first on each sample individually, and then on a combination of all samples. The resulting call sets were then merged using the CombineVariants module of GATK29 3.3.0.
Variant annotation was then performed using ANNOVAR30, cross-referencing the databases RefGene31, COSMIC32 v79 and dbSNP33 v138 with flagged somatic and clinically associated variants removed, ESP6500 (http://evs.gs.washington.edu/EVS), 1000 Genomes34 (August 2015). Synonymous variants, variants in non-exonic regions and those with an entry in the non-flagged dbSNP 138 database were then discarded. For increased specificity, only known COSMIC variants in oncogenes were retained. For tumor suppressors, non-COSMIC variants were also considered, provided that the variant was not present in the ESP6500 or 1000 Genomes databases. This was motivated by the fact that tumor suppressors are more likely to be subject to novel loss of function mutations. Additional manual inspection of variants was performed in IGV35, and novel mutations adjacent to homopolymer repeats or those only occurring at the ends of reads were discarded. Two additional potential mutations at known or recurrently mutated sites, although with very few supporting reads (2, 1, and 2 respectively), were added for samples M130111 (NRAS), M140602A (NRAS), and M120511B-2 (DDX3X) after visual inspection. The final set of mutations were visualized with Maftools36 R package.
Gene expression analysis
Differential expression analysis was performed in R, using the DESeq237 package (default parameters), after first filtering out the unexpressed genes. Batch effects were accounted for in the DESeq2 regression model, since the samples M130128A, M130204B, M141204, M150313, and M160212 were sequenced at a different date. To visualize gene expression values, normalized and batch corrected counts were derived using the “removeBatchEffect” function in the limma38 package, based on values obtained with the “variance stabilizing transformation” DESeq2 method. Gene set enrichment analyses were performed on gene lists ranked by the log2-fold changes estimated by DESeq2 using FGSEA39, based on the MSigDB40 “Canonical pathways” gene set. A minimum gene set size of 25 and a maximum of 500 was specified and 1 million permutations were demanded.
Cell lines
The melanoma cell lines SK-MEL-2 and MML-1 were purchased from the Cell Lines Service GmbH (Eppelheim, Germany), and HEK-293T was purchased from ATCC, Manassas, VA). They were cultured in 37 °C and 5% CO2 in the presence of DMEM-F12 (SK-MEL-2), RPMI1640 (MML-1), or DMEM media (HEK-293T) (Gibco), supplemented with 10% fetal bovine serum (Gibco) and Gentamycin (Thermo Fisher Scientific, Waltham, MA).
Transfection
MML-1 cells were transfected with a pool of five different siRNAs against DDX3X (SMARTpool, Dharmacon) in the presence of DhermaFECT (Dharmacon).
Cell cycle analysis
Harvested cells were re-suspended in modified Vindelövs solution (20 mM Tris, 100 mM NaCl, 1 µg/mL 7-aminoactinomycin D (7-AAD), 20 µg/mL RNase, and 0.1% nonidet P-40, adjusted to pH 8.0). The samples were analyzed using a BD Accuri C6 flow cytometer.
Quantification of ABCB1 protein expression in vitro
Harvested cells were fixed and permeabilized (Fixation/Permeabilization solution, BD Bioscience). Next, the cells were stained with monoclonal rabbit ABCB1 antibody (Cell Signaling #13342) followed by staining with a secondary anti-Rabbit FITC (DAKO F0205). The stained cells were analyzed using a BD Accuri C6 flow cytometer.
Analysis of p-gp pump activity
Cells were grown in the presence of 200 ng/mL Rhodamine 123 for 60 min with or without Elacridar (1 µM). After incubation, the cells were washed with PBS and cultured for another 90 min in fresh medium with or withour Elacridar (1 µM). Cell were harvested and re-suspended in PBS and analyzed with a BD Accuri C6 flow cytometer.
Flow cytometry sorting of ABCB1 expressing cells
Cells were cultured in the presence of Rhodamine 123 (200 ng/mL) for 60 min. The harvested cells were sorted based on negative Rhodamine 123 signal, using Facs Aria IIIµ.
Virus production and transduction
The calcium phosphate precipitation method was used to produce retroviruses by transfecting HEK-293T cells with the following plasmids; pHaMDRwt (kind gift from Michael Gottesman, Addgene plasmid #10957)41 and MSCV-IRES-PURO. Target cells were transduced over night and analyzed 72 h after transduction.
Degranulation assay
TILs (tumor infiltrating lymphocytes) were extracted, cultured, and expanded in vitro by culturing primary tumor pieces in the presence of IL2. Next, the TILs and autologous tumor cells were incubated separately overnight with 0.5 µM karonudib or DMSO (1:20000), after which 3*105 TILs and 1*105 tumor cells were plated together in a 96 well V-bottom plate and cultured with monoclonal antibody for CD107a (LAMP-1)-APC human (clone: REA792) (Milteny Biotech). After the 6-h incubation, cells were washed once and re-suspended in MACS buffer before the surface expression of CD107 was determined by flow cytometry using a BD Accuri C6 flow cytometer.
Measurement of IFN-gamma
IFN-gamma concentration in cell culture medium was measured using an enzyme-linked immunosorbent assay (ELISA) (Diaclone).
PDXv2 mouse model
The method of making PDXv2 mouse models has been previously described9. Briefly, patient-derived tumor sample (M151002) was transduced with a pHAGE-luc-GFP virus and transplanted subcutaneously on the flanks of the immune deficient mice. Once tumor growth was confirmed by bioluminescence measurements (PerkinElmer IVIS Lumina III XR), the mice were separated in four treatment groups of four mice receiving either karonudib or vehicle (same concentrations and dosing schedule as in the pre-clinical PDX trial) with or without TILs (one injection of 20*106 TILs). Tumor growth was followed by caliper and bioluminescence measurements.
Statistical analysis
Graphs and statistical testing were generated using GraphPad Prism, error bars on growth curves are shown as standard error of mean (SEM), and other error bars are shown as Standard error of mean (SD). Student’s t test was performed, where statistically significant P value was indicated as; *P < 0.05, **P < 0.01, ***P < 0.001, or ****P < 0.0001.
Change history
06 February 2020
On Pubmed, the name of co-author Roger Olofsson Bagge appeared incorrectly as “Bagge RO” instead of “Olofsson Bagge, Roger”. This has been corrected in the PDF and HTML versions.
References
Garbe, C. et al. Diagnosis and treatment of melanoma. European consensus-based interdisciplinary guideline - update 2016. Eur. J. Cancer 63, 201–217 (2016).
Ugurel, S. et al. Survival of patients with advanced metastatic melanoma: the impact of novel therapies-update 2017. Eur. J. Cancer 83, 247–257 (2017).
Gad, H. et al. MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool. Nature 508, 215–221 (2014).
Warpman Berglund, U. et al. Validation and development of MTH1 inhibitors for treatment of cancer. Ann. Oncol. 27, 2275–2283 (2016).
Kawamura, T. et al. Proteomic profiling of small-molecule inhibitors reveals dispensability of MTH1 for cancer cell survival. Sci. Rep. 6, 26521 (2016).
Kettle, J. G. et al. Potent and selective inhibitors of MTH1 probe its role in cancer cell survival. J. Med. Chem. 59, 2346–2361 (2016).
Wang, J. Y. et al. Reactive oxygen species dictate the apoptotic response of melanoma cells to TH588. J. Invest. Dermatol. 136, 2277–2286 (2016).
Einarsdottir, B. O. et al. Melanoma patient-derived xenografts accurately model the disease and develop fast enough to guide treatment decisions. Oncotarget 5, 9609–9618 (2014).
Jespersen, H. et al. Clinical responses to adoptive T-cell transfer can be modeled in an autologous immune-humanized mouse model. Nat. Commun. 8, 707 (2017).
Gao, H. et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat. Med. 21, 1318–1325 (2015).
Ben-David, U. et al. Patient-derived xenografts undergo mouse-specific tumor evolution. Nat. Genet. 49, 1567–1575 (2017).
Eisenhauer, E. A. et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur. J. Cancer 45, 228–247 (2009).
Patel, A. et al. MutT Homolog 1 (MTH1) maintains multiple KRAS-driven pro-malignant pathways. Oncogene 34, 2586–2596 (2015).
Rai, P. et al. Continuous elimination of oxidized nucleotides is necessary to prevent rapid onset of cellular senescence. Proc. Natl Acad. Sci. USA 106, 169–174 (2009).
Rai, P. et al. Enhanced elimination of oxidized guanine nucleotides inhibits oncogenic RAS-induced DNA damage and premature senescence. Oncogene 30, 1489–1496 (2011).
Valentin-Vega, Y. A. et al. Cancer-associated DDX3X mutations drive stress granule assembly and impair global translation. Sci. Rep. 6, 25996 (2016).
Chernov, K. G. et al. Role of microtubules in stress granule assembly: microtubule dynamical instability favors the formation of micrometric stress granules in cells. J. Biol. Chem. 284, 36569–36580 (2009).
Binkhathlan, Z. & Lavasanifar, A. P-glycoprotein inhibition as a therapeutic approach for overcoming multidrug resistance in cancer: current status and future perspectives. Curr. Cancer Drug Targets 13, 326–346 (2013).
Kunjachan, S., Rychlik, B., Storm, G., Kiessling, F. & Lammers, T. Multidrug resistance: physiological principles and nanomedical solutions. Adv. Drug Deliv. Rev. 65, 1852–1865 (2013).
Schinkel, A. H. et al. Disruption of the mouse mdr1a P-glycoprotein gene leads to a deficiency in the blood-brain barrier and to increased sensitivity to drugs. Cell 77, 491–502 (1994).
Leslie, E. M., Deeley, R. G. & Cole, S. P. Multidrug resistance proteins: role of P-glycoprotein, MRP1, MRP2, and BCRP (ABCG2) in tissue defense. Toxicol. Appl. Pharmacol. 204, 216–237 (2005).
Wilmott, J. S. et al. Selective BRAF inhibitors induce marked T-cell infiltration into human metastatic melanoma. Clin. Cancer Res. 18, 1386–1394 (2012).
Taube, J. M. et al. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin. Cancer Res. 20, 5064–5074 (2014).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Harrow, J. et al. GENCODE: the reference human genome annotation for the ENCODE project. Genome Res. 22, 1760–1774 (2012).
Mudge, J. M. & Harrow, J. Creating reference gene annotation for the mouse C57BL6/J genome assembly. Mamm. Genome 26, 366–378 (2015).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Koboldt, D. C. et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22, 568–576 (2012).
Warden, C. D., Adamson, A. W., Neuhausen, S. L. & Wu, X. Detailed comparison of two popular variant calling packages for exome and targeted exon studies. PeerJ 2, e600 (2014).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
O’Leary, N. A. et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745 (2016).
Forbes, Sa et al. COSMIC: mining complete cancer genomes in the catalogue of somatic mutations in cancer. Nucleic Acids Res. 39, 945–950 (2011).
Sherry, S. T. et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311 (2001).
The Genomes Project, C., Vol. 526 68–74 (2015).
Thorvaldsdóttir, H., Robinson, J. T. & Mesirov, J. P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 14, 178–192 (2013).
Mayakonda, A. & Koeffler, H. P. Maftools: efficient analysis, visualization and summarization of MAF files from large-scale cohort based cancer studies. bioRxiv https://doi.org/10.1101/052662 (2016).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–34 (2014).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Sergushichev, A. An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. bioRxiv https://doi.org/10.1101/060012 (2016).
Subramanian, A., et al., Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA, 2005. 102(43): p. 15545-50.
Pastan, I. et al. A retrovirus carrying an MDR1 cDNA confers multidrug resistance and polarized expression of P-glycoprotein in MDCK cells. Proc. Natl Acad. Sci. USA 85, 4486–4490 (1988).
Acknowledgements
We wish to thank Sofia Stenqvist for the assistance with animal experiments. This work was supported by the Swedish Cancer Society and the Swedish Research Council (to J.A.N., T.H., and E.L.), the Knut and Alice Wallenberg Foundation (to J.A.N. and T.H.) and the Region Västra Götaland (Sahlgrenska University Hospital, Gothenburg), the IngaBritt and Arne Lundberg Foundation, BioCARE—a National Strategic Cancer Research Program at University of Gothenburg, the Lion’s Cancer Foundation West and Familjen Erling Perssons Stiftelse (to J.A.N.), the Swedish Foundation for Strategic Research (T.H.), the Assar Gabrielsson Foundation, the W&M Lundgren Foundation and Sahlgrenska Universitetssjukhusets stiftelser (Sahlgrenska University Hospital, Gothenburg) (to B.O.E., E.M.V.S. and M.F.L.), and the European Academy of Dermatology and Venereology and the Fondation Nuovo Soldati (to E.F.-B.).
Author information
Authors and Affiliations
Contributions
B.O.E., J.K., E.M.V.S., M.F.L., S.F.B., E.F.-B., and H.J. performed the experiments. J.K. did the bioinformatics. R.O.B., L.C., U.S., and L.N. recruited patients and supplied patient biopsies. E.L., L.M.N., and J.A.N. supervised the research. M.S., T.K., O.W., U.W.B., and T.H. synthesized and supplied karonudib and contributed the know-how. B.O.E. and J.K. generated the figures and B.O.E. and J.N. wrote the paper, but all authors edited and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
M.S., T.K., O.W., U.W.B., T.H., and J.A.N. are shareholders of Oxcia, which commercialize anti-cancer drug development projects, including MTH1 inhibitors from the Helleday Foundation. The remaining authors declare that they have no conflict of interest.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by G. Giaccone
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Einarsdottir, B.O., Karlsson, J., Söderberg, E.M. et al. A patient-derived xenograft pre-clinical trial reveals treatment responses and a resistance mechanism to karonudib in metastatic melanoma. Cell Death Dis 9, 810 (2018). https://doi.org/10.1038/s41419-018-0865-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-018-0865-6
This article is cited by
-
MTH1 as a target to alleviate T cell driven diseases by selective suppression of activated T cells
Cell Death & Differentiation (2022)
-
A multicenter clinical study: personalized medication for advanced gastrointestinal carcinomas with the guidance of patient-derived tumor xenograft (PDTX)
Journal of Cancer Research and Clinical Oncology (2022)
-
Karonudib has potent anti-tumor effects in preclinical models of B-cell lymphoma
Scientific Reports (2021)
-
circRNA-miRNA-mRNA regulatory network in human lung cancer: an update
Cancer Cell International (2020)
-
Applications of patient-derived tumor xenograft models and tumor organoids
Journal of Hematology & Oncology (2020)