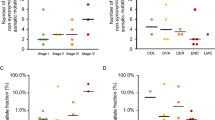
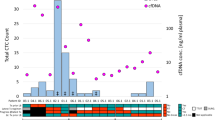
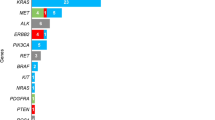
Abstract
Liquid biopsy, a minimally invasive approach for detecting tumor biomarkers in blood, has emerged as a leading-edge technique in cancer precision medicine. New evidence has shown that liquid biopsies can incidentally detect pathogenic germline variants (PGVs) associated with cancer predisposition, including in patients with a cancer for which genetic testing is not recommended. The ability to detect these incidental PGV in cancer patients through liquid biopsy raises important questions regarding the management of this information and its clinical implications. This incidental identification of PGVs raises concerns about cancer predisposition and the potential impact on patient management, not only in terms of providing access to treatment based on the tumor molecular profiling, but also the management of revealing genetic predisposition in patients and families. Understanding how to interpret this information is essential to ensure proper decision-making and to optimize cancer treatment and prevention strategies. In this review we provide a comprehensive summary of current evidence of incidental PGVs in cancer predisposition genes identified by liquid biopsy in patients with cancer. We critically review the methodological considerations of liquid biopsy as a tool for germline diagnosis, clinical utility and potential implications for cancer prevention, treatment, and research.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Malapelle U, Tiseo M, Vivancos A, Kapp J, Serrano MJ, Tiemann M. Liquid biopsy for biomarker testing in non-small cell lung cancer: a European perspective. J Mol Pathol. 2021;2:255–73.
Wan JCM, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17:223–38.
Sidransky D. Nucleic acid-based methods for the detection of cancer. Science. 1997;278:1054–9.
Thierry AR, El Messaoudi S, Gahan PB, Anker P, Stroun M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 2016;35:347–76.
Haber DA, Velculescu VE. Blood-based analyses of cancer: circulating tumor cells and circulating tumor DNA. Cancer Discov. 2014;4:650–61.
Fernández-Lázaro D, García Hernández JL, García AC, Córdova Martínez A, Mielgo-Ayuso J, Cruz-Hernández JJ. Liquid biopsy as novel tool in precision medicine: origins, properties, identification and clinical perspective of cancer’s biomarkers. Diagnostics (Basel). 2020;10:215.
Heitzer E, Haque IS, Roberts CES, Speicher MR. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet. 2019;20:71–88.
Garcia EP, Minkovsky A, Jia Y, Ducar MD, Shivdasani P, Gong X, et al. Validation of OncoPanel: a targeted next-generation sequencing assay for the detection of somatic variants in cancer. Arch Pathol Lab Med. 2017;141:751–8.
Lone SN, Nisar S, Masoodi T, Singh M, Rizwan A, Hashem S, et al. Liquid biopsy: a step closer to transform diagnosis, prognosis and future of cancer treatments. Mol Cancer. 2022;21:79.
Elazezy M, Joosse SA. Techniques of using circulating tumor DNA as a liquid biopsy component in cancer management. Comput Struct Biotechnol J. 2018;16:370–8.
Bohers E, Viailly PJ, Jardin F. cfDNA sequencing: technological approaches and bioinformatic issues. Pharm (Basel). 2021;14:596.
Gezer U, Bronkhorst AJ, Holdenrieder S. The clinical utility of droplet digital PCR for profiling circulating tumor DNA in breast cancer patients. Diagnostics (Basel). 2022;12:3042.
Babayan A, Pantel K. Advances in liquid biopsy approaches for early detection and monitoring of cancer. Genome Med. 2018;10:21.
Willis J, Lefterova MI, Artyomenko A, Kasi PM, Nakamura Y, Mody K, et al. Validation of microsatellite instability detection using a comprehensive plasma-based genotyping panel. Clin Cancer Res. 2019;25:7035–45.
Kim ST, Cristescu R, Bass AJ, Kim KM, Odegaard JI, Kim K, et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat Med. 2018;24:1449–58.
Garcia-Murillas I, Schiavon G, Weigelt B, Ng C, Hrebien S, Cutts RJ, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med. 2015;7:302ra133.
Tie J, Wang Y, Tomasetti C, Li L, Springer S, Kinde I, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016;8:346ra92.
Chaudhuri AA, Chabon JJ, Lovejoy AF, Newman AM, Stehr H, Azad TD, et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov. 2017;7:1394–403.
Adashek JJ, Janku F, Kurzrock R. Signed in blood: circulating tumor DNA in cancer diagnosis, treatment and screening. Cancers (Basel). 2021;13:3600.
Hu Y, Ulrich BC, Supplee J, Kuang Y, Lizotte PH, Feeney NB, et al. False-positive plasma genotyping due to clonal hematopoiesis. Clin Cancer Res. 2018;24:4437–43.
Aldea M, Tagliamento M, Bayle A, Vasseur D, Vergé V, Marinello A, et al. Liquid biopsies for circulating tumor DNA detection may reveal occult hematologic malignancies in patients with solid tumors. JCO Precis Oncol. 2023;7:e2200583.
Slavin TP, Banks K, Chudova D, Oxnard GR, Odegaard JI, Nagy RJ, et al. Identification of putative germline mutations in 10,288 patients undergoing circulating tumor DNA testing. JCO. 2017;35:1514–1514.
Lui YYN, Chik KW, Chiu RWK, Ho CY, Lam CWK, Lo YMD. Predominant hematopoietic origin of cell-free DNA in plasma and serum after sex-mismatched bone marrow transplantation. Clin Chem. 2002;48:421–7.
Barbitoff YA, Abasov R, Tvorogova VE, Glotov AS, Predeus AV. Systematic benchmark of state-of-the-art variant calling pipelines identifies major factors affecting accuracy of coding sequence variant discovery. BMC Genomics. 2022;23:155.
Stout LA, Kassem N, Hunter C, Philips S, Radovich M, Schneider BP. Identification of germline cancer predisposition variants during clinical ctDNA testing. Sci Rep. 2021;11:13624.
Strom SP. Current practices and guidelines for clinical next-generation sequencing oncology testing. Cancer Biol Med. 2016;13:3–11.
Odegaard JI, Vincent JJ, Mortimer S, Vowles JV, Ulrich BC, Banks KC, et al. Validation of a plasma-based comprehensive cancer genotyping assay utilizing orthogonal tissue- and plasma-based methodologies. Clin Cancer Res. 2018;24:3539–49.
Bando H, Nakamura Y, Taniguchi H, Shiozawa M, Yasui H, Esaki T, et al. Effects of metastatic sites on circulating tumor DNA in patients with metastatic colorectal cancer. JCO Precis Oncol. 2022;6:e2100535.
Mandelker D, Donoghue M, Talukdar S, Bandlamudi C, Srinivasan P, Vivek M, et al. Germline-focussed analysis of tumour-only sequencing: recommendations from the ESMO Precision Medicine Working Group. Ann Oncol. 2019;30:1221–31.
Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–7.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Mersch J, Brown N, Pirzadeh-Miller S, Mundt E, Cox HC, Brown K, et al. Prevalence of variant reclassification following hereditary cancer genetic testing. JAMA. 2018;320:1266–74.
Slavin TP, Banks KC, Chudova D, Oxnard GR, Odegaard JI, Nagy RJ, et al. Identification of incidental germline mutations in patients with advanced solid tumors who underwent cell-free circulating tumor DNA sequencing. J Clin Oncol. 2018;36:JCO1800328.
Pascual J, Attard G, Bidard FC, Curigliano G, De Mattos-Arruda L, Diehn M, et al. ESMO recommendations on the use of circulating tumour DNA assays for patients with cancer: a report from the ESMO Precision Medicine Working Group. Ann Oncol. 2022;33:750–68.
Lanman RB, Mortimer SA, Zill OA, Sebisanovic D, Lopez R, Blau S, et al. Analytical and clinical validation of a digital sequencing panel for quantitative, highly accurate evaluation of cell-free circulating tumor DNA. PLoS One. 2015;10:e0140712.
Woodhouse R, Li M, Hughes J, Delfosse D, Skoletsky J, Ma P, et al. Clinical and analytical validation of FoundationOne Liquid CDx, a novel 324-Gene cfDNA-based comprehensive genomic profiling assay for cancers of solid tumor origin. PLoS One. 2020;15:e0237802.
Kuzbari Z, Bandlamudi C, Loveday C, Garrett A, Mehine M, George A, et al. Germline-focused analysis of tumour-detected variants in 49,264 cancer patients: ESMO Precision Medicine Working Group recommendations. Ann Oncol. 2023;34:215–27.
Vogelstein B, Kinzler KW. The multistep nature of cancer. Trends Genet. 1993;9:138–41.
Bunz F. Principles of Cancer Genetics [Internet]. Dordrecht: Springer Netherlands; [cited 2023 Jul 11]. Available from: http://link.springer.com/10.1007/978-94-017-7484-0 (2016).
Daly MB, Pal T, Berry MP, Buys SS, Dickson P, Domchek SM, et al. Genetic/familial high-risk assessment: breast, ovarian, and pancreatic, version 2.2021, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2021;19:77–102.
Weiss JM, Gupta S, Burke CA, Axell L, Chen LM, Chung DC, et al. NCCN Guidelines® insights: genetic/familial high-risk assessment: colorectal, version 1.2021. J Natl Compr Canc Netw. 2021;19:1122–32.
Armstrong N, Ryder S, Forbes C, Ross J, Quek RG. A systematic review of the international prevalence of BRCA mutation in breast cancer. Clin Epidemiol. 2019;11:543–61.
Pritchard CC, Mateo J, Walsh MF, De Sarkar N, Abida W, Beltran H, et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N. Engl J Med. 2016;375:443–53.
Tung N, Dougherty KC, Gatof ES, DeLeonardis K, Hogan L, Tukachinsky H, et al. Potential pathogenic germline variant reporting from tumor comprehensive genomic profiling complements classic approaches to germline testing. NPJ Precis Oncol. 2023;7:76.
Mezquita L, Bucheit LA, Laguna JC, Pastor B, Teixido C, Gorria T, et al. Prevalence of incidental pathogenic germline variants detected in cfDNA in patients with oncogene-driven non-small cell lung cancer. JCO. 2022;40:10569–10569.
Mezquita L, Bucheit L, Laguna JC, Pastor B, Teixido C, Gorria T, et al. MA07.07 clinical/molecular profile of patients with non-small cell lung cancer (NSCLC) with incidental pathogenic germline variants detected in cfDNA. J Thorac Oncol. 2022;17:S69.
Chang CM, Lin KC, Hsiao NE, Hong WA, Lin CY, Liu TC, et al. Clinical application of liquid biopsy in cancer patients. BMC Cancer. 2022;22:413.
Ratajska M, Koczkowska M, Żuk M, Gorczyński A, Kuźniacka A, Stukan M, et al. Detection of BRCA1/2 mutations in circulating tumor DNA from patients with ovarian cancer. Oncotarget. 2017;8:101325–32.
Hu Y, Alden RS, Odegaard JI, Fairclough SR, Chen R, Heng J, et al. Discrimination of Germline EGFR T790M Mutations in Plasma Cell-Free DNA Allows Study of Prevalence Across 31,414 Cancer Patients. Clin Cancer Res. 2017;23:7351–9.
Vidula N, Rich TA, Sartor O, Yen J, Hardin A, Nance T, et al. Routine plasma-based genotyping to comprehensively detect germline, somatic, and reversion BRCA mutations among patients with advanced solid tumors. Clin Cancer Res. 2020;26:2546–55.
Liu M, Niu X, Liu H, Chen J. Germline EGFR mutations in lung cancer (Review). Oncol Lett. 2023;26:282.
Shukuya T, Patel S, Shane-Carson K, He K, Bertino EM, Shilo K, et al. Lung cancer patients with germline mutations detected by next-generation sequencing and/or liquid biopsy. J Thorac Oncol. 2018;13:e17–9.
Lakhani SR, Van De Vijver MJ, Jacquemier J, Anderson TJ, Osin PP, McGuffog L, et al. The pathology of familial breast cancer: predictive value of immunohistochemical markers estrogen receptor, progesterone receptor, HER-2, and p53 in patients with mutations in BRCA1 and BRCA2. J Clin Oncol. 2002;20:2310–8.
Chen H, Wu J, Zhang Z, Tang Y, Li X, Liu S, et al. Association between BRCA status and triple-negative breast cancer: a meta-analysis. Front Pharm. 2018;9:909.
Imai K, Yamamoto H. Carcinogenesis and microsatellite instability: the interrelationship between genetics and epigenetics. Carcinogenesis. 2008;29:673–80.
Kuba MG, Lester SC, Bowman T, Stokes SM, Taneja KL, Garber JE, et al. Histopathologic features of breast cancer in Li-Fraumeni syndrome. Mod Pathol. 2021;34:542–8.
NCCN [Internet]. [cited 2023 Apr 1]. Detection, Prevention, and Risk Reduction. Available from: https://www.nccn.org/guidelines/category_2.
Hampel H, Bennett RL, Buchanan A, Pearlman R, Wiesner GL, Guideline Development Group, American College of Medical Genetics and Genomics Professional Practice and Guidelines Committee and National Society of Genetic Counselors Practice Guidelines Committee. A practice guideline from the American College of Medical Genetics and Genomics and the National Society of Genetic Counselors: referral indications for cancer predisposition assessment. Genet Med. 2015;17:70–87.
Belleau P, Deschênes A, Chambwe N, Tuveson DA, Krasnitz A. Genetic ancestry inference from cancer-derived molecular data across genomic and transcriptomic platforms. Cancer Res. 2023;83:49–58.
Prakash R, Zhang Y, Feng W, Jasin M. Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb Perspect Biol. 2015;7:a016600.
Robson M, Im SA, Senkus E, Xu B, Domchek SM, Masuda N, et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med. 2017;377:523–33.
Litton JK, Rugo HS, Ettl J, Hurvitz SA, Gonçalves A, Lee KH, et al. Talazoparib in patients with advanced breast cancer and a germline BRCA mutation. N Engl J Med. 2018;379:753–63.
Domchek SM, Aghajanian C, Shapira-Frommer R, Schmutzler RK, Audeh MW, Friedlander M, et al. Efficacy and safety of olaparib monotherapy in germline BRCA1/2 mutation carriers with advanced ovarian cancer and three or more lines of prior therapy. Gynecol Oncol. 2016;140:199–203.
Oza AM, Tinker AV, Oaknin A, Shapira-Frommer R, McNeish IA, Swisher EM, et al. Antitumor activity and safety of the PARP inhibitor rucaparib in patients with high-grade ovarian carcinoma and a germline or somatic BRCA1 or BRCA2 mutation: Integrated analysis of data from Study 10 and ARIEL2. Gynecol Oncol. 2017;147:267–75.
Moore K, Colombo N, Scambia G, Kim BG, Oaknin A, Friedlander M, et al. Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med. 2018;379:2495–505.
Ray-Coquard I, Pautier P, Pignata S, Pérol D, González-Martín A, Berger R, et al. Olaparib plus bevacizumab as first-line maintenance in ovarian cancer. N Engl J Med. 2019;381:2416–28.
González-Martín A, Pothuri B, Vergote I, DePont Christensen R, Graybill W, Mirza MR, et al. Niraparib in patients with newly diagnosed advanced ovarian cancer. N. Engl J Med. 2019;381:2391–402.
Golan T, Hammel P, Reni M, Van Cutsem E, Macarulla T, Hall MJ, et al. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N Engl J Med. 2019;381:317–27.
de Bono J, Mateo J, Fizazi K, Saad F, Shore N, Sandhu S, et al. Olaparib for metastatic castration-resistant prostate cancer. N Engl J Med. 2020;382:2091–102.
Abida W, Patnaik A, Campbell D, Shapiro J, Bryce AH, McDermott R, et al. Rucaparib in men with metastatic castration-resistant prostate cancer harboring a BRCA1 or BRCA2 gene alteration. J Clin Oncol. 2020;38:3763–72.
Mylavarapu S, Das A, Roy M. Role of BRCA mutations in the modulation of response to platinum therapy. Front Oncol. 2018;8:16.
Vencken PMLH, Kriege M, Hoogwerf D, Beugelink S, van der Burg MEL, Hooning MJ, et al. Chemosensitivity and outcome of BRCA1- and BRCA2-associated ovarian cancer patients after first-line chemotherapy compared with sporadic ovarian cancer patients. Ann Oncol. 2011;22:1346–52.
Emelyanova M, Pudova E, Khomich D, Krasnov G, Popova A, Abramov I, et al. Platinum-based chemotherapy for pancreatic cancer: impact of mutations in the homologous recombination repair and Fanconi anemia genes. Ther Adv Med Oncol. 2022;14:17588359221083050.
Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–20.
Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409–13.
André T, Shiu KK, Kim TW, Jensen BV, Jensen LH, Punt C, et al. Pembrolizumab in microsatellite-instability-high advanced colorectal cancer. N Engl J Med. 2020;383:2207–18.
Marabelle A, Le DT, Ascierto PA, Di Giacomo AM, De Jesus-Acosta A, Delord JP, et al. Efficacy of pembrolizumab in patients with noncolorectal high microsatellite instability/mismatch repair-deficient cancer: results from the phase II KEYNOTE-158 study. J Clin Oncol. 2020;38:1–10.
Thariat J, Chevalier F, Orbach D, Ollivier L, Marcy PY, Corradini N, et al. Avoidance or adaptation of radiotherapy in patients with cancer with Li-Fraumeni and heritable TP53-related cancer syndromes. Lancet Oncol. 2021;22:e562–74.
Ballinger ML, Ferris NJ, Moodie K, Mitchell G, Shanley S, James PA, et al. Surveillance in Germline TP53 Mutation Carriers Utilizing Whole-Body Magnetic Resonance Imaging. JAMA Oncol. 2017;3:1735–6.
Stjepanovic N, Moreira L, Carneiro F, Balaguer F, Cervantes A, Balmaña J, et al. Hereditary gastrointestinal cancers: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. † Ann Oncol 2019;30:1558–71.
Sessa C, Balmaña J, Bober SL, Cardoso MJ, Colombo N, Curigliano G, et al. Risk reduction and screening of cancer in hereditary breast-ovarian cancer syndromes: ESMO Clinical Practice Guideline. Ann Oncol. 2023;34:33–47.
Rebbeck TR, Friebel T, Lynch HT, Neuhausen SL, van ’t Veer L, Garber JE, et al. Bilateral prophylactic mastectomy reduces breast cancer risk in BRCA1 and BRCA2 mutation carriers: the PROSE Study Group. J Clin Oncol. 2004;22:1055–62.
Domchek SM, Friebel TM, Singer CF, Evans DG, Lynch HT, Isaacs C, et al. Association of risk-reducing surgery in BRCA1 or BRCA2 mutation carriers with cancer risk and mortality. JAMA. 2010;304:967–75.
Whitaker KD, Obeid E, Daly MB, Hall MJ. Cascade genetic testing for hereditary cancer risk: an underutilized tool for cancer prevention. JCO Precis Oncol. 2021;5:1387–96.
Veyseh M, Ricker C, Espenschied C, Raymond V, D’Souza A, Barzi A. Secondary germline finding in liquid biopsy of a deceased patient; case report and review of the literature. Front Oncol. 2018;8:259.
Hunter CL, Helft PR. Yes, we can, but should we? ethical considerations in reporting germline findings from paired tumor-normal genomic testing in patients with advanced cancer. J Clin Oncol. 2023;41:1982–5.
Rolfo C, Cardona AF, Cristofanilli M, Paz-Ares L, Diaz Mochon JJ, Duran I, et al. Challenges and opportunities of cfDNA analysis implementation in clinical practice: Perspective of the International Society of Liquid Biopsy (ISLB). Crit Rev Oncol Hematol. 2020;151:102978.
Behel V, Noronha V, Choughule A, Shetty O, Chandrani P, Kapoor A, et al. Impact of molecular tumor board on the clinical management of patients with cancer. JCO Glob Oncol. 2022;8:e2200030.
Acknowledgements
We thank Ainara Arcocha, Jessica González and Laura Alcolea for the administrative support and Sarah Mackenzie PhD, for language editing.
Funding
The authors received no specific funding for this work. Juan Carlos Laguna received support from Contractes Clínic de Recerca “Emili Letang-Josep Font” 2023; Hospital Clínic Barcelona, 2023. Laura Mezquita received support from the Contrato Juan Rodes 2020 (ISCIII, Ministry of Health; JR20/00019); Ayuda de la Acción Estratégica en Salud- ISCIII FIS 2021 (PI21/01653); Ayuda SEOM Juan Rodés 2020 and Beca SEOM Grupo emergente 2022.
Author information
Authors and Affiliations
Contributions
JCL, BP and LM: Conceived and designed the review. All: Drafted and reviewed the manuscript. All: Approved the final version.
Corresponding author
Ethics declarations
Competing interests
JCL: Lectures and educational activities: Kyowa Kirin; Travel, Accommodations, Expenses: Rovi, Pierre-Fabre. BP: The author declares no conflict of interest. IN: The author declares no conflict of interest. SH: The author declares no conflict of interest. CT: Lectures and educational activities: AstraZeneca, Roche, Janssen Oncology, Pfizer, Biocartis, Merck Sharp & Dohme; Consulting, advisory role: Novartis, AstraZeneca; Research Grants: Novartis, AstraZeneca; Travel, Accommodations, Expenses: Bristol-Myers Squibb, Lilly, Merck Sharp & Dohme, Pfizer. MP: Educational activities: ISDIN. JAP: The author declares no conflict of interest. LM: Lectures and educational activities: Bristol-Myers Squibb, AstraZeneca, Roche, Takeda, Janssen, Pfizer, MSD; Consulting, advisory role: Roche, Takeda, Janssen; Research Grants: Bristol-Myers Squibb, Boehringer Ingelheim, Amgen, Stilla, Inivata, AstraZeneca; Travel, Accommodations, Expenses: Bristol-Myers Squibb, Roche, Takeda, AstraZeneca, Janssen.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Laguna, J.C., Pastor, B., Nalda, I. et al. Incidental pathogenic germline alterations detected through liquid biopsy in patients with solid tumors: prevalence, clinical utility and implications. Br J Cancer (2024). https://doi.org/10.1038/s41416-024-02607-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41416-024-02607-9