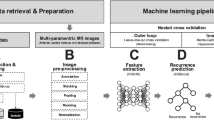
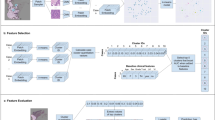
Abstract
Background
Accurate estimation of the long-term risk of recurrence in patients with non-metastatic colorectal cancer (CRC) is crucial for clinical management. Histology-based deep learning is expected to provide more abundant information for risk stratification.
Methods
We developed and validated a weakly supervised deep-learning model for predicting 5-year relapse-free survival (RFS) to stratify patients with different risks based on histological images from three hospitals of 614 cases with non-metastatic CRC. A deep prognostic factor (DL-RRS) was established to stratify patients into high and low-risk group. The areas under the curve (AUCs) were calculated to evaluate the performances of models.
Results
Our proposed model achieves the AUCs of 0.833 (95% CI: 0.736–0.905) and 0.715 (95% CI: 0.647–0.776) on validation cohort and external test cohort, respectively. The 5-year RFS rate was 45.7% for high DL-RRS patients, and 82.5% for low DL-RRS patients respectively in the external test cohort (HR: 3.89, 95% CI: 2.51–6.03, P < 0.001). Adjuvant chemotherapy was associated with improved RFS in Stage II patients with high DL-RRS (HR: 0.15, 95% CI: 0.06–0.38, P < 0.001).
Conclusions
DL-RRS has a good predictive performance of 5-year recurrence risk in CRC, and will better serve the clinical decision-making.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The data from the First Affiliated Hospital of Sun Yat-sen University, the Sun Yat-sen University Cancer Center and Shunde Hospital of Southern Medical University that support the findings of this study are available upon reasonable request from the corresponding author (SP). The data are not publicly available because they contain protected patient privacy information. Authors will share deidentified individual participant imaging data on request with researchers who provide a methodologically viable proposal and can do analyses that achieve the aims of the proposal. To gain access to data, requestors will need to sign a data access agreement.
Code availability
The training code base for the deep-learning framework is available at https://github.com/PRAETORIANCOHORT/Predicting_5_year_Recurrence_Risk_in_Colorectal_Cancer_a_Histology-Based_Deep_Learning_Approach.
References
André T, Boni C, Mounedji-Boudiaf L, Navarro M, Tabernero J, Hickish T, et al. Oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment for colon cancer. New Engl J Med. 2004;350:2343–51.
Kuebler JP, Wieand HS, O’Connell MJ, Smith RE, Colangelo LH, Yothers G, et al. Oxaliplatin combined with weekly bolus fluorouracil and leucovorin as surgical adjuvant chemotherapy for stage II and III colon cancer: results from NSABP C-07. J Clin Oncol. 2007;25:2198–204.
André T, de Gramont A, Vernerey D, Chibaudel B, Bonnetain F, Tijeras-Raballand A, et al. Adjuvant fluorouracil, leucovorin, and oxaliplatin in stage II to III colon cancer: updated 10-year survival and outcomes according to BRAF mutation and mismatch repair status of the MOSAIC study. J Clin Oncol. 2015;33:4176–87.
Grothey A, Sobrero AF, Shields AF, Yoshino T, Paul J, Taieb J, et al. Duration of adjuvant chemotherapy for stage III colon cancer. New Engl J Med. 2018;378:1177–88.
Amin MB, Greene FL, Edge SB, Compton CC, Gershenwald JE, Brookland RK, et al. The Eighth Edition AJCC Cancer Staging Manual: continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J Clin. 2017;67:93–99.
Gunderson LL, Jessup JM, Sargent DJ, Greene FL, Stewart AK. Revised TN categorization for colon cancer based on national survival outcomes data. J Clin Oncol. 2010;28:264–71.
O’Connell JB, Maggard MA, Ko CY. Colon cancer survival rates with the new American Joint Committee on Cancer sixth edition staging. J Natl Cancer Inst. 2004;96:1420–5.
Kim MJ, Jeong SY, Choi SJ, Ryoo SB, Park JW, Park KJ, et al. Survival paradox between stage IIB/C (T4N0) and stage IIIA (T1-2N1) colon cancer. Ann Surg Oncol. 2015;22:505–12.
Weiser MR, Gönen M, Chou JF, Kattan MW, Schrag D. Predicting survival after curative colectomy for cancer: individualizing colon cancer staging. J Clin Oncol. 2011;29:4796–802.
Renfro LA, Grothey A, Xue Y, Saltz LB, André T, Twelves C, et al. ACCENT-based web calculators to predict recurrence and overall survival in stage III colon cancer. J Natl Cancer Inst. 2014;106:dju333.
Sjoquist KM, Renfro LA, Simes RJ, Tebbutt NC, Clarke S, Seymour MT, et al. Personalizing survival predictions in advanced colorectal cancer: the ARCAD Nomogram Project. J Natl Cancer Inst. 2018;110:638–48.
Venook AP, Niedzwiecki D, Lopatin M, Ye X, Lee M, Friedman PN, et al. Biologic determinants of tumor recurrence in stage II colon cancer: validation study of the 12-gene recurrence score in cancer and leukemia group B (CALGB) 9581. J Clin Oncol. 2013;31:1775–81.
Zhang JX, Song W, Chen ZH, Wei JH, Liao YJ, Lei J, et al. Prognostic and predictive value of a microRNA signature in stage II colon cancer: a microRNA expression analysis. Lancet Oncol. 2013;14:1295–306.
Niedzwiecki D, Frankel WL, Venook AP, Ye X, Friedman PN, Goldberg RM, et al. Association between results of a gene expression signature assay and recurrence-free interval in patients with stage II colon cancer in cancer and leukemia group B 9581 (Alliance). J Clin Oncol. 2016;34:3047–53.
Yamanaka T, Oki E, Yamazaki K, Yamaguchi K, Muro K, Uetake H, et al. 12-Gene recurrence score assay stratifies the recurrence risk in stage II/III colon cancer with surgery alone: the SUNRISE study. J Clin Oncol. 2016;34:2906–13.
Pagès F, Mlecnik B, Marliot F, Bindea G, Ou FS, Bifulco C, et al. International validation of the consensus Immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet. 2018;391:2128–39.
Pagès F, André T, Taieb J, Vernerey D, Henriques J, Borg C, et al. Prognostic and predictive value of the Immunoscore in stage III colon cancer patients treated with oxaliplatin in the prospective IDEA France PRODIGE-GERCOR cohort study. Ann Oncol. 2010;31:921–9.
Lugli A, Zlobec I, Berger MD, Kirsch R, Nagtegaal ID. Tumour budding in solid cancers. Nat Rev Clin Oncol. 2021;18:101–15.
Niazi MKK, Parwani AV, Gurcan MN. Digital pathology and artificial intelligence. Lancet Oncol. 2019;20:e253–e261.
Lu MY, Chen TY, Williamson DFK, Zhao M, Shady M, Lipkova J, et al. AI-based pathology predicts origins for cancers of unknown primary. Nature. 2021;594:106–10.
Kulkarni PM, Robinson EJ, Sarin Pradhan J, Gartrell-Corrado RD, Rohr BR, Trager MH, et al. Deep learning based on standard H&E images of primary melanoma tumors identifies patients at risk for visceral recurrence and death. Clin Cancer Res. 2020;26:1126–34.
Saillard C, Schmauch B, Laifa O, Moarii M, Toldo S, Zaslavskiy M, et al. Predicting survival after hepatocellular carcinoma resection using deep learning on histological slides. Hepatology. 2020;72:2000–13.
Woerl AC, Eckstein M, Geiger J, Wagner DC, Daher T, Stenzel P, et al. Deep learning predicts molecular subtype of muscle-invasive bladder cancer from conventional histopathological slides. Eur Urol. 2020;78:256–64.
Sirinukunwattana K, Domingo E, Richman SD, Redmond KL, Blake A, Verrill C, et al. Image-based consensus molecular subtype (imCMS) classification of colorectal cancer using deep learning. Gut. 2021;70:544–54.
Yamashita R, Long J, Longacre T, Peng L, Berry G, Martin B, et al. Deep learning model for the prediction of microsatellite instability in colorectal cancer: a diagnostic study. Lancet Oncol. 2021;22:132–41.
Kather JN, Pearson AT, Halama N, Jäger D, Krause J, Loosen SH, et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat Med. 2019;25:1054–6.
Skrede OJ, De Raedt S, Kleppe A, Hveem TS, Liestøl K, Maddison J, et al. Deep learning for prediction of colorectal cancer outcome: a discovery and validation study. Lancet. 2020;395:350–60.
Renehan AG, Egger M, Saunders MP, O’Dwyer ST. Impact on survival of intensive follow up after curative resection for colorectal cancer: systematic review and meta-analysis of randomised trials. BMJ. 2002;324:813.
Tjandra JJ, Chan MK. Follow-up after curative resection of colorectal cancer: a meta-analysis. Dis Colon Rectum. 2007;50:1783–99.
Seo SI, Lim SB, Yoon YS, Kim CW, Yu CS, Kim TW, et al. Comparison of recurrence patterns between ≤5 years and >5 years after curative operations in colorectal cancer patients. J Surg Oncol. 2013;108:9–13.
Benson AB, Venook AP, Al-Hawary MM, Arain MA, Chen YJ, Ciombor KK, et al. Colon cancer, version 2.2021, NCCN clinical practice guidelines in oncology. J Natl Compr Cancer Netw. 2021;19:329–59.
Zhao JJ, Lu DH, Ma K, Zhang Y, Zheng YF. Deep image clustering with category-style representation. In: Proceedings of the Computer Vision–ECCV 2020: 16th European Conference, Glasgow, UK, August 23–28, 2020, Proceedings, Part XIV 16, F, 2020.
Di Narzo AF, Tejpar S, Rossi S, Yan P, Popovici V, Wirapati P, et al. Test of four colon cancer risk-scores in formalin fixed paraffin embedded microarray gene expression data. J Natl Cancer Inst. 2014;106:dju247.
Hur K, Toiyama Y, Okugawa Y, Ide S, Imaoka H, Boland CR, et al. Circulating microRNA-203 predicts prognosis and metastasis in human colorectal cancer. Gut. 2017;66:654–65.
Reichling C, Taieb J, Derangere V, Klopfenstein Q, Le Malicot K, Gornet JM, et al. Artificial intelligence-guided tissue analysis combined with immune infiltrate assessment predicts stage III colon cancer outcomes in PETACC08 study. Gut. 2020;69:681–90.
Sirinukunwattana K, Snead D, Epstein D, Aftab Z, Mujeeb I, Tsang YW, et al. Novel digital signatures of tissue phenotypes for predicting distant metastasis in colorectal cancer. Sci Rep. 2018;8:13692.
Geessink OGF, Baidoshvili A, Klaase JM, Ehteshami Bejnordi B, Litjens GJS, van Pelt GW, et al. Computer aided quantification of intratumoral stroma yields an independent prognosticator in rectal cancer. Cell Oncol. 2019;42:331–41.
Quasar Collaborative Group, Gray R, Barnwell J, McConkey C, Hills RK, Williams NS, et al. Adjuvant chemotherapy versus observation in patients with colorectal cancer: a randomised study. Lancet. 2007;370:2020–9.
André T, Boni C, Navarro M, Tabernero J, Hickish T, Topham C, et al. Improved overall survival with oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment in stage II or III colon cancer in the MOSAIC trial. J Clin Oncol. 2009;27:3109–16.
Shi JY, Wang X, Ding GY, Dong Z, Han J, Guan Z, et al. Exploring prognostic indicators in the pathological images of hepatocellular carcinoma based on deep learning. Gut. 2021;70:951–61.
Maoz A, Dennis M, Greenson JK. The Crohn’s-like lymphoid reaction to colorectal cancer-tertiary lymphoid structures with immunologic and potentially therapeutic relevance in colorectal cancer. Front Immunol. 2019;10:1884.
Schumacher TN, Thommen DS. Tertiary lymphoid structures in cancer. Science. 2022;375:eabf9419.
West NP, Dattani M, McShane P, Hutchins G, Grabsch J, Mueller W, et al. The proportion of tumour cells is an independent predictor for survival in colorectal cancer patients. Br J Cancer. 2010;102:1519–23.
Huijbers A, Tollenaar RA, v Pelt GW, Zeestraten EC, Dutton S, McConkey CC, et al. The proportion of tumor-stroma as a strong prognosticator for stage II and III colon cancer patients: validation in the VICTOR trial. Ann Oncol. 2013;24:179–85.
Rozek LS, Schmit SL, Greenson JK, Tomsho LP, Rennert HS, Rennert G, et al. Tumor-infiltrating lymphocytes, Crohn’s-like lymphoid reaction, and survival from colorectal cancer. J Natl Cancer Inst. 2016;108:djw027.
Acknowledgements
We would like to thank the participants of the study and all the study staff for their contributions to the study.
Funding
This study was funded by grants from the National Key Research and Development Program of China (2020AAA0109504), the National Natural Science Foundation of China (82270557, 82272942 and 82203642), the Guangdong Natural Science Foundation (2021A1515010252), and the Guangzhou Science and Technology Plan Project (2023A04J2212).
Author information
Authors and Affiliations
Contributions
XS, HX and SP supervised the study. MK, XS, HX and SP conceived of and designed the study. WW and ZW trained and developed the artificial intelligence model. WW and BL did the statistical analysis. WW, ZW, SC and HX wrote the drafted report. YS, MK, XS, HX and SP critically revised the manuscript. WW, ZW, SC, JS, JL and KS organised and screened patients. All authors had access to all the raw datasets. XS, HX and SP verified all the data. All authors revised the report and approved the final version before submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This study protocol was centrally approved by the institutional Clinical Research Ethics Committee of the First Affiliated Hospital of Sun Yat-sen University (FAHSYSU), which conforms to the ethical guidelines of the 1975 Declaration of Helsinki. As this was a retrospective cohort study, informed consent was waived.
Consent for publication
Not applicable.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xiao, H., Weng, Z., Sun, K. et al. Predicting 5-year recurrence risk in colorectal cancer: development and validation of a histology-based deep learning approach. Br J Cancer 130, 951–960 (2024). https://doi.org/10.1038/s41416-024-02573-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-024-02573-2