Abstract
Background
Available data on Mismatch Repair system (MMR) deficiency are conflicting and derived from small studies. Our study aimed to evaluate the therapeutic implications of MMR status in patients with locally advanced rectal cancer (LARC).
Methods
We retrospectively collected data from 318 patients affected by LARC treated in Italy at the Medical Oncology Units of the University Hospital of Cagliari, Istituto Nazionale dei Tumori Milan, and AOU Ospedali Riuniti Ancona. All patients underwent neoadjuvant chemoradiotherapy. The primary objective was major TRG while secondary objectives were pathological complete response, disease-free survival (DFS) and overall survival (OS).
Results
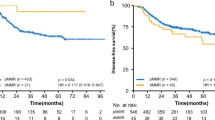
One hundred sixty patients (148 pMMR and 12 dMMR) were included in the exploratory cohort and 158 (146 pMMR and 12 dMMR) were included in the validation cohort. A major TRG has been shown in 42.6% and 43.1% patients with pMMR in exploratory and validation cohort, respectively; while no major TRG have been shown in dMMR patients in both cohorts. Exploratory and validation cohorts showed a statistically significant higher mDFS in pMMR patients compared to dMMR: NR vs. 14 months and NR vs. 17 months, respectively.
Conclusion
Our results indicated an association between dMMR and poor response to preoperative chemoradiotherapy and they represent a hypothesis-generating data for new neoadjuvant strategies.

This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
Datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. https://doi.org/10.3322/caac.21492.
Glynne-Jones R, Wyrwicz L, Tiret E, Brown G, Rödel C, Cervantes A, et al. Rectal cancer: ESMO clinical practice guidelines for diagnosis, treatment, and follow-up. Ann Oncol. 2017;28:iv22–40. https://doi.org/10.1093/annonc/mdx224.
Bosset JF, Calais G, Mineur L, Maingon P, Stojanovic-Rundic S, Bensadounet RJ, EORTC Radiation Oncology Group, et al. Fluorouracil-based adjuvant chemotherapy after preoperative chemoradiotherapy in rectal cancer: long-term results of the EORTC 22921 randomized study. Lancet Oncol. 2014;15:184–90. https://doi.org/10.1016/S1470-2045(13)70599-0.
Weiser MR. Total neoadjuvant therapy for locally advanced rectal cancer: PRODIGE 23 trial. Ann Surg Oncol. 2022;29:1493–5. https://doi.org/10.1245/s10434-021-11104-9.
Bahadoer RR, Dijkstra EA, van Etten B, Marijnen CAM, Putter H, Meershoek-Klein Kranenbarg E, et al. Short-course radiotherapy followed by chemotherapy before total mesorectal excision (TME) versus preoperative chemoradiotherapy, TME, and optional adjuvant chemotherapy in locally advanced rectal cancer (RAPIDO): a randomized, open-label, phase 3 trial. Lancet Oncol. 2021;22:29–42. https://doi.org/10.1016/S1470-2045(20)30555-6.
Garcia-Aguilar J, Patil S, Kim JK, Yuval JJB, Thompson H, Verheijet F, et al. Preliminary results of the organ preservation of rectal adenocarcinoma (OPRA) trial. J Clin Oncol. 2020;38:4008a. https://doi.org/10.1200/JCO.2020.38.15_suppl.4008.
Giunta EF, Bregni G, Pretta A, Deleporte A, Liberale G, Bali AM, et al. Total neoadjuvant therapy for rectal cancer: making sense of the results from the RAPIDO and PRODIGE 23 trials. Cancer Treat Rev. 2021;96:102177. https://doi.org/10.1016/j.ctrv.2021.102177.
Bregni G, Vandeputte C, Pretta A, Senti C, Trevisi E, Acedo Reina E, et al. Rationale and design of REGINA, a phase II trial of neoadjuvant regorafenib, nivolumab, and short-course radiotherapy in stage II and III rectal cancer. Acta Oncol. 2021;60:549–53. https://doi.org/10.1080/0284186X.2020.1871067.
Gutierrez ME, Price KS, Lanman RB, Nagy RJ, Shah I, Mathura S, et al. Genomic profiling for KRAS, NRAS, BRAF, microsatellite instability, and mismatch repair deficiency among patients with metastatic colon cancer. JCO Precis Oncol. 2019;3:PO.19.00274. https://doi.org/10.1200/PO.19.00274.
Ziranu P, Lai E, Schirripa M, Puzzoni M, Persano M, Pretta A, et al. The role of p53 expression in patients with RAS/BRAF wild-type metastatic colorectal cancer receiving irinotecan and cetuximab as later line treatment. Target Oncol. 2021;16:517–27. https://doi.org/10.1007/s11523-021-00816-3.
Vega-Benedetti AF, Loi E, Moi L, Restivo A, Cabras F, Deidda S, et al. Colorectal cancer promoter methylation alteration affects the expression of glutamate ionotropic receptor AMPA type subunit 4 alternative isoforms potentially relevant in colon tissue. Hum Cell. 2022;35:310–9. https://doi.org/10.1007/s13577-021-00640-x.
Giampieri R, Ziranu P, Daniele B, Zizzi A, Ferrari D, Lonardi S, et al. From CENTRAL to SENTRAL (SErum aNgiogenesis cenTRAL): circulating predictive biomarkers to anti-VEGFR therapy. Cancers. 2020;12:1330. https://doi.org/10.3390/cancers12051330.
Giampieri R, Lupi A, Ziranu P, Bittoni A, Pretta A, Pecci F, et al. Retrospective comparative analysis of KRAS G12C vs. other KRAS mutations in mCRC patients treated with first-line chemotherapy doublet + bevacizumab. Front Oncol. 2021;11:736104. https://doi.org/10.3389/fonc.2021.736104.
Lai E, Liscia N, Donisi C, Mariani S, Tolu S, Pretta A, et al. Molecular-biology-driven treatment for metastatic colorectal cancer. Cancers. 2020;12:1214. https://doi.org/10.3390/cancers12051214.
Pasqualetti G, Schirripa M, Dochy E, Fassan M, Ziranu P, Puzzoni M, et al. Thyroid hormones ratio is a major prognostic marker in advanced metastatic colorectal cancer: results from the phase III randomised CORRECT trial. Eur J Cancer. 2020;133:66–73. https://doi.org/10.1016/j.ejca.2020.04.023.
Puzzoni M, Ziranu P, Demurtas L, Lai E, Mariani S, Liscia N, et al. Why precision medicine should be applied across the continuum of care for metastatic colorectal cancer patients. Future Oncol. 2020;16:4337–9. https://doi.org/10.2217/fon-2019-0624.
Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA. 1998;95:6870–5. https://doi.org/10.1073/pnas.95.12.6870.
Timothy MP, Chandrajit PR, Rodriguez-Bigas MA. Colorectal carcinogenesis: MSI-H versus MSI-L. Dis Markers. 2004;20:199–206. https://doi.org/10.1155/2004/368680.
Tran B, Kopetz S, Tie J, Gibbs P, Jiang ZQ, Lieu CH, et al. Impact of BRAF mutation and microsatellite instability on the pattern of metastatic spread and prognosis in metastatic colorectal cancer. Cancer. 2011;117:4623–32. https://doi.org/10.1002/cncr.26086.
Merok MA, Ahlquist T, Royrvik EC, Tufteland KF, Hektoen M, Sjo OH, et al. Microsatellite instability has a positive prognostic impact on stage II colorectal cancer after complete resection: results from a large, consecutive Norwegian series. Ann Oncol. 2013;24:1274–82. https://doi.org/10.1093/annonc/mds614.
Sargent DJ, Marsoni S, Monges G, Thibodeau SN, Labianca R, Hamilton SR, et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol. 2010;28:3219–26. https://doi.org/10.1200/JCO.2009.27.1825.
Sargent DJ, Marsoni S, Thibodeau SN, Labianca R, Hamilton SR, Torri V, et al. Confirmation of deficient mismatch repair (dMMR) as a predictive marker for lack of benefit from 5-FU based chemotherapy in stage II and III colon cancer (CC): a pooled molecular reanalysis of randomized chemotherapy trials. J Clin Oncol. 2008;26:15s. https://doi.org/10.1200/jco.2008.26.15_suppl.4008.
Brueckl WM, Moesch C, Brabletz T, Koebnick C, Riedel C, Jung A, et al. Relationship between microsatellite instability, response and survival in palliative patients with colorectal cancer undergoing first-line chemotherapy. Anticancer Res. 2003;23:1773–7.
Alex AK, Siqueira S, Coudry R, Santos J, Alves M, Hoff PM, et al. Response to chemotherapy and prognosis in metastatic colorectal cancer with DNA deficient mismatch repair. Clin Colorectal Cancer. 2017;16:228–39. https://doi.org/10.1016/j.clcc.2016.11.001.
Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–20. https://doi.org/10.1056/NEJMoa1500596.
Overman MJ, Lonardi S, Wong KYM, Lenz HJ, Gelsomino F, Aglietta M, et al. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair—deficient/microsatellite instability—high metastatic colorectal cancer. JCO. 2018;36:773–9. https://doi.org/10.1200/JCO.2017.76.9901.
Overman MJ, Lonardi S, Wong KYM, Lenz HJ, Gelsomino F, Aglietta M, et al. Nivolumab (NIVO) + low-dose ipilimumab (IPI) in previously treated patients (pts) with microsatellite instability-high/mismatch repair-deficient (MSI-H/dMMR) metastatic colorectal cancer (mCRC): long-term follow-up. JCO. 2019;37:635. https://doi.org/10.1200/JCO.2019.37.4_suppl.635.
Andre T, Shiu KK, Kim TW, Jensen BV, Jensen LH, Punt CJA, et al. Pembrolizumab versus chemotherapy for microsatellite instability-high/mismatch repair deficient metastatic colorectal cancer: the phase 3 KEYNOTE-177 study. In: Proceedings of the ASCO Annual Meeting 2020, Virtual Scientific Program, Chicago, IL, USA, 29–31 May 2020. https://doi.org/10.1200/JCO.2020.38.18_suppl.LBA4.
Becht E, de Reyniès A, Giraldo NA, Pilati C, Buttard B, Lacroix L, et al. Immune and stromal classification of colorectal cancer is associated with molecular subtypes and relevant for precision immunotherapy. Clin Cancer Res. 2016;22:4057–66. https://doi.org/10.1158/1078-0432.CCR-15-2879.
Giannakis M, Mu XJ, Shukla SA, Qian ZR, Cohen O, Nishihara R, et al. Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep. 2016;15:857–65. https://doi.org/10.1016/j.celrep.2016.03.075.
Salem ME, Bodor JN, Puccini A, Xiu J, Goldberg RM, Grothey A, et al. Relationship between MLH1, PMS2, MSH2, and MSH6 gene-specific alterations and tumor mutational burden in 1057 microsatellite instability-high solid tumors. Int J Cancer. 2020. https://doi.org/10.1002/ijc.33115.
Benson AB III, Venook AP, Cederquist L, Chan E, Chen YJ, Cooper HS, et al. Colon cancer, version 1.2017, NCCN clinical practice guidelines in oncology. J Natl Compr Cancer Netw. 2017;15:370–98. https://doi.org/10.6004/jnccn.2017.0036.
de Rosa N, Rodriguez-Bigas MA, Chang GJ, Veerapong J, Borras E, Krishnan S, et al. DNA mismatch repair deficiency in rectal cancer: benchmarking its impact on prognosis, neoadjuvant response prediction, and clinical cancer genetics. J Clin Oncol. 2016;34:3039–46. https://doi.org/10.1200/JCO.2016.66.6826.
Cercek A, Dos Santos Fernandes G, Roxburgh CS, Ganesh K, Ng S, Sanchez-Vega F, et al. Mismatch repair-deficient rectal cancer and resistance to neoadjuvant chemotherapy. Clin Cancer Res. 2020;26:3271–9. https://doi.org/10.1158/1078-0432.CCR-19-3728.
Choi YY, Kim H, Shin SJ, Kim HY, Lee J, Yang HK, et al. Microsatellite instability and programmed cell death-ligand 1 expression in stage II/III gastric cancer: post hoc analysis of the CLASSIC randomized controlled study. Ann Surg. 2019;270:309–16. https://doi.org/10.1097/SLA.0000000000002803.
Smyth EC, Wotherspoon A, Peckitt C, Gonzalez D, Hulkki-Wilson S, Eltahir Z, et al. Mismatch repair deficiency, microsatellite instability, and survival: an exploratory analysis of the medical research council adjuvant gastric infusional chemotherapy (MAGIC) trial. JAMA Oncol. 2017;3:1197–203. https://doi.org/10.1001/jamaoncol.2016.6762.
Pietrantonio F, Miceli R, Raimondi A, Kim YW, Kang WK, Langley RE, et al. Individual patient data meta-analysis of the value of microsatellite instability as a biomarker in gastric cancer. J Clin Oncol. 2019;37:3392–3400. https://doi.org/10.1200/JCO.19.01124.
Ryan R, Gibbons D, Hyland JM, Treanor D, White A, Mulcahy HE, et al. Pathological response following long-course neoadjuvant chemoradiotherapy for locally advanced rectal cancer. Histopathology. 2005;47:141–6. https://doi.org/10.1111/j.1365-2559.2005.02176.x.
Huh JW, Kim HC, Kim SH, Park YA, Cho YN, Yun SH, et al. Tumor regression grade as a clinically useful outcome predictor in patients with rectal cancer after preoperative chemoradiotherapy. Surgery. 2019;165:579–85. https://doi.org/10.1016/j.surg.2018.08.026.
Jäger T, Neureiter D, Urbas R, Klieser E, Hitzl W, Emmanuelet K. Applicability of American Joint Committee on cancer and College of American Pathologists Regression Grading System in rectal cancer. Dis Colon Rectum. 2017;60:815–26. https://doi.org/10.1097/DCR.0000000000000806.
Dworak O, Keilholz L, Hoffmann A. Pathological features of rectal cancer after preoperative radiochemotherapy. Int J Colorectal Dis. 1997;12:19–23. https://doi.org/10.1007/s003840050072.
Cercek A, Lumish M, Sinopoli J, Weiss J, Shia J, Lamendola-Essel M, et al. PD-1 blockade in mismatch repair-deficient, locally advanced rectal cancer. N Engl J Med. 2022;386:2363–76. https://doi.org/10.1056/NEJMoa2201445.
Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335–46. https://doi.org/10.1038/nrm1907.
Palombo F, Gallinari P, Iaccarino I, Lettieri T, Hughes M, D’Arrigo A, et al. GTBP, a 160-kilodalton protein essential for mismatch-binding activity in human cells. Science. 1995;268:1912–4. https://doi.org/10.1126/science.7604265.
Drummond JT, Li GM, Longley MJ, Modrich P. Isolation of an hMSH2–p160 heterodimer that restores DNA mismatch repair to tumor cells. Science. 1995;268:1909–12. https://doi.org/10.1126/science.7604264.
Acharya S, Wilson T, Gradia S, Kane MF, Guerrette S, Marsischky GT, et al. hMSH2 forms specific mispairbinding complexes with hMSH3 and hMSH6. Proc Natl Acad Sci USA. 1996;93:13629–34. https://doi.org/10.1073/pnas.93.24.13629.
Palombo F, Iaccarino I, Nakajima E, Ikejima M, Shimada T, Jiricnyet J. hMutSβ, a heterodimer of hMSH2 and hMSH3, binds to insertion/deletion loops in DNA. Curr Biol. 1996;6:1181–4. https://doi.org/10.1016/s0960-9822(02)70685-4.
Gradia S, Acharya S, Fishel R. The human mismatch recognition complex hMSH2–hMSH6 functions as a novel molecular switch. Cell. 1997;91:995–1005. https://doi.org/10.1016/s0092-8674(00)80490-0.
Prolla TA, Baker SM, Harris AC, Tsao JL, Yao X, Bronneret CE, et al. Tumour susceptibility and spontaneous mutation in mice deficient in Mlh1, Pms1 and Pms2 DNA mismatch repair. Nature Genet. 1998;18:276–9. https://doi.org/10.1038/ng0398-276.
Raschle M, Marra G, Nystrom-Lahti M, Schar P, Jiricny J. Identification of hMutLβ, a heterodimer of hMLH1 and hPMS1. J Biol Chem. 1999;274:32368–75. https://doi.org/10.1074/jbc.274.45.32368.
Chen PC, Dudley S, Hagen W, Dizon D, Paxton L, Reichow D, et al. Contributions by MutL homologue Mlh3 and Pms2 to DNA mismatch repair and tumor suppression in the mouse. Cancer Res. 2005;65:8662–70. https://doi.org/10.1158/0008-5472.CAN-05-0742.
Cannavo E, Marra G, Sabates-Bellver J, Menigatti M, Lipkin SM, Fischeret F, et al. Expression of the MutL homologue hMLH3 in human cells and its role in DNA mismatch repair. Cancer Res. 2005;65:10759–66. https://doi.org/10.1158/0008-5472.CAN-05-2528.
Flores-Rozas H, Kolodner RD. The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frameshift mutations. Proc Natl Acad Sci USA. 1998;95:12404–9. https://doi.org/10.1073/pnas.95.21.12404.
Stojic L, Brun R, Jiricny J. Mismatch repair, and DNA damage signaling. DNA Repair. 2004;3:1091–101. https://doi.org/10.1016/j.dnarep.2004.06.006.
Duckett DR, Drummond JT, Murchie AI, Reardon JT, Sancar A, Lilley DM, et al. Human MutSα recognizes damaged DNA base pairs containing O6-methylguanine, O4-methylthymine, or the cisplatin(GpG) adduct. Proc Natl Acad Sci USA. 1996;93:6443–7. https://doi.org/10.1073/pnas.93.13.6443.
Karran P. Mechanisms of tolerance to DNA damaging therapeutic drugs. Carcinogenesis. 2001;22:1931–7. https://doi.org/10.1093/carcin/22.12.1931.
Cheah PL, Li J, Looi LM, Koh CC, Lau TP, Chang SW, et al. Screening for microsatellite instability in colorectal carcinoma: practical utility of immunohistochemistry and PCR with fragment analysis in a diagnostic histopathology setting. Malays J Pathol. 2019;41:91–100.
Hissong E, Crowe EP, Yantiss RK, Chen YT. Assessing colorectal cancer mismatch repair status in the modern era: a survey of current practices and re-evaluation of the role of microsatellite instability testing. Mod Pathol. 2018;31:1756–66. https://doi.org/10.1038/s41379-018-0094-7.
Shia J, Klimstra DS, Nafa K, Offit K, Guillem JG, Markowitz AJ, et al. Value of immunohistochemical detection of DNA mismatch repair proteins in predicting germline mutation in hereditary colorectal neoplasms. Am J Surg Pathol. 2005;29:96–104. https://doi.org/10.1097/01.pas.0000146009.85309.3b.
Baretti M, Le DT. DNA mismatch repair in cancer. Pharmacol Ther. 2018;189:45–62.
Author information
Authors and Affiliations
Contributions
AP: conceptualization, methodology, validation, formal analysis, investigation, resources, data curation, writing—original draft, writing—review and editing, visualization. PZ and RG: resources, data curation, formal analysis, writing—original draft, writing—review and editing. G Pinna and CD: resources, data curation, writing—original draft. GR, FL, GD, EP, FM and FS: resources, data curation. AR, SM, MAD, VP, MP, EL, AR, LZ, R Barbara, and R Berardi: resources. G Pretta and CS: writing—review and editing. GF: resources, data curation, writing—original draft, writing—review and editing. FP: resources, data curation, writing—original draft, writing—review and editing. MS: methodology, validation, formal analysis, investigation, resources, data curation, writing—original draft, writing—review and editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Ethics Committee approval was obtained for the study (Protocol number 2020/10912—code: EMIBIOCCOR) from Cagliari Independent Ethics Committee and written informed consent was obtained from all participants for their tissues to be utilized for this work. This study was performed in accordance with the study protocol, the ethical principles stated in the Declaration of Helsinki as well as those indicated in the International Conference on Harmonization (ICH) Note for Guidance on Good Clinical Practice (GCP; ICH E6, 1995), and all applicable regulatory requirements. All patients signed a written informed consent before study entry. Adequate information was given to eligible patients by the principal investigator or co-investigators in accordance with local regulations. The declaration of informed consent was personally signed and dated by the subject, and by the investigator/person designated by the investigator to conduct the informed consent discussion.
Consent for publication
Patients signed an informed consent regarding the publication of their data.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pretta, A., Ziranu, P., Giampieri, R. et al. Mismatch Repair system protein deficiency as a resistance factor for locally advanced rectal adenocarcinoma patients receiving neoadjuvant chemo-radiotherapy. Br J Cancer 129, 1619–1624 (2023). https://doi.org/10.1038/s41416-023-02444-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-023-02444-2