Abstract
Background
Delta-like ligand 3 (DLL3) is a therapeutic target in small-cell lung cancer (SCLC). However, how DLL3 expression status affects the tumor microenvironment (TME) and clinical outcomes in SCLC remains unclear.
Methods
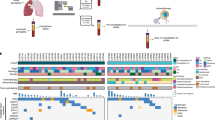
This retrospective study included patients with postoperative limited-stage (LS)-SCLC and extensive-stage (ES)-SCLC treated with platinum and etoposide (PE) plus anti-programmed cell death ligand 1 (PD-L1) antibody. We investigated the relationship of DLL3 expression with TME, mutation status, tumor neoantigens, and immunochemotherapy.
Results
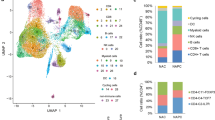
In the LS-SCLC cohort (n = 59), whole-exome sequencing revealed that DLL3High cases had significantly more neoantigens (P = 0.004) and a significantly higher rate of the signature SBS4 associated with smoking (P = 0.02) than DLL3Low cases. Transcriptome analysis in the LS-SCLC cohort revealed that DLL3High cases had significantly suppressed immune-related pathways and dendritic cell (DC) function. SCLC with DLL3High had significantly lower proportions of T cells, macrophages, and DCs than those with DLL3Low. In the ES-SCLC cohort (n = 30), the progression-free survival associated with PE plus anti-PD-L1 antibody was significantly worse in DLL3High cases than in DLL3Low cases (4.7 vs. 7.4 months, P = 0.01).
Conclusions
Although SCLC with DLL3High had a higher neoantigen load, these tumors were resistant to immunochemotherapy due to suppressed tumor immunity by inhibiting antigen-presenting functions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during this study are available from the corresponding author on reasonable request.
Change history
03 November 2023
A Correction to this paper has been published: https://doi.org/10.1038/s41416-023-02481-x
References
Gazdar AF, Bunn PA, Minna JD. Small-cell lung cancer: what we know, what we need to know and the path forward. Nat Rev Cancer. 2017;17:725–37. https://doi.org/10.1038/nrc.2017.87.
Horn L, Mansfield AS, Szczesna A, Havel L, Krzakowski M, Hochmair MJ, et al. First-line atezolizumab plus chemotherapy in extensive-stage small-cell lung cancer. N Engl J Med. 2018;379:2220–9. https://doi.org/10.1056/NEJMoa1809064.
Paz-Ares L, Dvorkin M, Chen Y, Reinmuth N, Hotta K, Trukhin D, et al. Durvalumab plus platinum–etoposide versus platinum–etoposide in first-line treatment of extensive-stage small-cell lung cancer (CASPIAN): a randomised, controlled, open-label, phase 3 trial. Lancet. 2019;394:1929–39. https://doi.org/10.1016/S0140-6736(19)32222-6.
Tanaka K, Isse K, Fujihira T, Takenoyama M, Saunders L, Bheddah S, et al. Prevalence of Delta-like protein 3 expression in patients with small cell lung cancer. Lung Cancer. 2018;115:116–20. https://doi.org/10.1016/j.lungcan.2017.11.018.
Morgensztern D, Besse B, Greillier L, Santana-Davila R, Ready N, Hann CL, et al. Efficacy and safety of rovalpituzumab tesirine in third-line and beyond patients with DLL3-expressing, relapsed/refractory small-cell lung cancer: results from the phase II TRINITY study. Clin Cancer Res. 2019;25:6958–66. https://doi.org/10.1158/1078-0432.Ccr-19-1133.
Baine MK, Hsieh MS, Lai WV, Egger JV, Jungbluth AA, Daneshbod Y, et al. SCLC subtypes defined by ASCL1, NEUROD1, POU2F3, and YAP1: a comprehensive immunohistochemical and histopathologic characterization. J Thorac Oncol. 2020;15:1823–35. https://doi.org/10.1016/j.jtho.2020.09.009.
Chapman G, Sparrow DB, Kremmer E, Dunwoodie SL. Notch inhibition by the ligand Delta-like 3 defines the mechanism of abnormal vertebral segmentation in spondylocostal dysostosis. Hum Mol Genet. 2011;20:905–16. https://doi.org/10.1093/hmg/ddq529.
Furuta M, Kikuchi H, Shoji T, Takashima Y, Kikuchi E, Kikuchi J, et al. DLL3 regulates the migration and invasion of small cell lung cancer by modulating Snail. Cancer Sc. 2019;110:1599–608. https://doi.org/10.1111/cas.13997.
Jiang T, Collins BJ, Jin N, Watkins DN, Brock MV, Matsui W, et al. Achaete-scute complex homologue 1 regulates tumor-initiating capacity in human small cell lung cancer. Cancer Res. 2009;69:845–54. https://doi.org/10.1158/0008-5472.Can-08-2762.
Henke RM, Meredith DM, Borromeo MD, Savage TK, Johnson JE. Ascl1 and Neurog2 form novel complexes and regulate Delta-like3 (Dll3) expression in the neural tube. Dev Biol. 2009;328:529–40. https://doi.org/10.1016/j.ydbio.2009.01.007.
Rudin CM, Poirier JT, Byers LA, Dive C, Dowlati A, George J, et al. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat Rev Cancer. 2019;19:289–97. https://doi.org/10.1038/s41568-019-0133-9.
Rudin CM, Pietanza MC, Bauer TM, Ready N, Morgensztern D, Glisson BS, et al. Rovalpituzumab tesirine, a DLL3-targeted antibody-drug conjugate, in recurrent small-cell lung cancer: a first-in-human, first-in-class, open-label, phase 1 study. Lancet Oncol. 2017;18:42–51.
Blackhall F, Jao K, Greillier L, Cho BC, Penkov K, Reguart N, et al. Efficacy and safety of rovalpituzumab tesirine compared with topotecan as second-line therapy in DLL3-high SCLC: results from the phase 3 TAHOE study. J Thorac Oncol. 2021;16:1547–58. https://doi.org/10.1016/j.jtho.2021.02.009.
Johnson ML, Zvirbule Z, Laktionov K, Helland A, Cho BC, Gutierrez V, et al. Rovalpituzumab tesirine as a maintenance therapy after first-line platinum-based chemotherapy in patients with extensive-stage–SCLC: results from the phase 3 MERU study. J Thorac Oncol. 2021;16:1570–81. https://doi.org/10.1016/j.jtho.2021.03.012.
Molloy ME, Valenzuela LB, Basak PE, Law CL. Abstract 1573: Combinatorial antitumor effects of CD3-based trispecific T cell activating constructs (TriTACs) and checkpoint inhibitors in preclinical models. Cancer Res. 2021;81:1573 https://doi.org/10.1158/1538-7445.Am2021-1573.
Giffin MJ, Cooke K, Lobenhofer EK, Estrada J, Zhan J, Deegen P, et al. AMG 757, a half-life extended, DLL3-targeted bispecific T-cell engager, shows high potency and sensitivity in preclinical models of small-cell lung cancer. Clin Cancer Res. 2021;27:1526–37. https://doi.org/10.1158/1078-0432.Ccr-20-2845.
Johnson ML, Sanborn RE. Interim results of an ongoing phase 1/2a study of HPN328, a tri-specific, half-life extended, DLL3-targeting, T-cell engager, in patients with small cell lung cancer and other neuroendocrine cancers. Headache. 2022;3:17.
Goldstraw P, Chansky K, Crowley J, Rami-Porta R, Asamura H, Eberhardt WE, et al. The IASLC lung cancer staging project: proposals for revision of the TNM stage groupings in the forthcoming (eighth) edition of the TNM classification for lung cancer. J Thorac Oncol. 2016;11:39–51. https://doi.org/10.1016/j.jtho.2015.09.009.
Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45:228–47. https://doi.org/10.1016/j.ejca.2008.10.026.
Fedchenko N, Reifenrath J. Different approaches for interpretation and reporting of immunohistochemistry analysis results in the bone tissue—a review. Diagn Pathol. 2014;9:221 https://doi.org/10.1186/s13000-014-0221-9.
Saito M, Shimada Y, Shiraishi K, Sakamoto H, Tsuta K, Totsuka H, et al. Development of lung adenocarcinomas with exclusive dependence on oncogene fusions. Cancer Res. 2015;75:2264–71. https://doi.org/10.1158/0008-5472.Can-14-3282.
Honda T, Sakashita H, Masai K, Totsuka H, Motoi N, Kobayashi M, et al. Deleterious pulmonary surfactant system gene mutations in lung adenocarcinomas associated with usual interstitial pneumonia. JCO Precis Oncol. 2018;2:1–24. https://doi.org/10.1200/po.17.00301.
DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8. https://doi.org/10.1038/ng.806.
Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–92. https://doi.org/10.1093/bib/bbs017.
Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. https://doi.org/10.1038/nbt.1754.
Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, Wu Y, et al. The repertoire of mutational signatures in human cancer. Nature. 2020;578:94–101. https://doi.org/10.1038/s41586-020-1943-3.
Huang X, Wojtowicz D, Przytycka TM. Detecting presence of mutational signatures in cancer with confidence. Bioinformatics. 2018;34:330–7. https://doi.org/10.1093/bioinformatics/btx604.
Hundal J, Carreno BM, Petti AA, Linette GP, Griffith OL, Mardis ER, et al. pVAC-Seq: a genome-guided in silico approach to identifying tumor neoantigens. Genome Med. 2016;8:11.
Hoof I, Peters B, Sidney J, Pedersen LE, Sette A, Lund O, et al. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics. 2009;61:1–13.
McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, et al. The Ensembl variant effect predictor. Genome Biol. 2016;17:122.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. https://doi.org/10.1093/bioinformatics/bts635.
Frankish A, Diekhans M, Ferreira AM, Johnson R, Jungreis I, Loveland J, et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47:D766–D773. https://doi.org/10.1093/nar/gky955.
Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290–5. https://doi.org/10.1038/nbt.3122.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50. https://doi.org/10.1073/pnas.0506580102.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550 https://doi.org/10.1186/s13059-014-0550-8.
Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, et al. IFN-γ–related mRNA profile predicts clinical response to PD-1 blockade. J Clin Investig. 2017;127:2930–40. https://doi.org/10.1172/JCI91190.
Coppola D, Nebozhyn M, Khalil F, Dai H, Yeatman T, Loboda A, et al. Unique ectopic lymph node-like structures present in human primary colorectal carcinoma are identified by immune gene array profiling. Am J Pathol. 2011;179:37–45. https://doi.org/10.1016/j.ajpath.2011.03.007.
Newman AM, Steen CB, Liu CL, Gentles AJ, Chaudhuri AA, Scherer F, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. 2019;37:773–82. https://doi.org/10.1038/s41587-019-0114-2.
Mahadevan NR, Knelson EH, Wolff JO, Vajdi A, Saigí M, Campisi M, et al. Intrinsic immunogenicity of small cell lung carcinoma revealed by its cellular plasticity. Cancer Discov. 2021;11:1952–69. https://doi.org/10.1158/2159-8290.Cd-20-0913.
Hubaux R, Thu KL, Coe BP, MacAulay C, Lam S, Lam WL. EZH2 promotes E2F-driven SCLC tumorigenesis through modulation of apoptosis and cell-cycle regulation. J Thorac Oncol. 2013;8:1102–6. https://doi.org/10.1097/JTO.0b013e318298762f.
Sen T, Rodriguez BL, Chen L, Corte CMD, Morikawa N, Fujimoto J, et al. Targeting DNA damage response promotes antitumor immunity through STING-mediated T-cell activation in small cell lung cancer. Cancer Discov. 2019;9:646–61. https://doi.org/10.1158/2159-8290.Cd-18-1020.
Saunders LR, Bankovich AJ, Anderson WC, Aujay MA, Bheddah S, Black K, et al. A DLL3-targeted antibody-drug conjugate eradicates high-grade pulmonary neuroendocrine tumor-initiating cells in vivo. Sci Transl Med. 2015;7:302ra136 https://doi.org/10.1126/scitranslmed.aac9459.
Augustyn A, Borromeo M, Wang T, Fujimoto J, Shao C, Dospoy PD, et al. ASCL1 is a lineage oncogene providing therapeutic targets for high-grade neuroendocrine lung cancers. Proc Natl Acad Sci USA. 2014;111:14788–93. https://doi.org/10.1073/pnas.1410419111.
Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348:69–74. https://doi.org/10.1126/science.aaa4971.
Newell F, Pires da Silva I, Johansson PA, Menzies AM, Wilmott JS, Addala V, et al. Multiomic profiling of checkpoint inhibitor-treated melanoma: identifying predictors of response and resistance, and markers of biological discordance. Cancer Cell. 2022;40:88–102.e7. https://doi.org/10.1016/j.ccell.2021.11.012.
Goldman JW, Garassino MC, Chen Y, Reinmuth N, Hotta K, Poltoratskiy A, et al. LBA86 Durvalumab (D) ± tremelimumab (T) + platinum-etoposide (EP) in 1L ES-SCLC: characterization of long-term clinical benefit and tumour mutational burden (TMB) in CASPIAN. Ann Oncol. 2020;31:S1212–S1213. https://doi.org/10.1016/j.annonc.2020.08.2328.
Slaney CY, Wang P, Darcy PK, Kershaw MH. CARs versus BiTEs: a comparison between T cell–redirection strategies for cancer treatment. Cancer Discov. 2018;8:924–34. https://doi.org/10.1158/2159-8290.Cd-18-0297.
Paz-Ares L, Champiat S, Lai WV, Izumi H, Govindan R, Boyer M, et al. Tarlatamab, a first-in-class DLL3-targeted bispecific T cell engager, in recurrent small-cell lung cancer: an open-label, phase 1 study. J Clin Oncol. 2023;41:2893–903. https://doi.org/10.1200/jco.22.02823.
Acknowledgements
We thank all patients who contributed to this study. We also thank all the investigators from the oncologic department of the National Cancer Center Hospital for their contributions.
Funding
This study was supported in part by the Grant for Lung Cancer Research from MEXT kakenhi Grant-in-Aid for Scientific Research (C) (Grant number 23K07615), the Japan Lung Cancer Society (TY), AMED (22hk0102080h0002), MEXT kakenhi Grant-in-Aid for Scientific Research (C) (Grant number 21K06900) (NM), JST AIP-PRISM (Grant Number JPMJCR18Y4), and MHLW ICT infrastructure establishment and implementation of artificial intelligence for the clinical and medical research program (Grant Number JP21AC5001) (RH).
Author information
Authors and Affiliations
Contributions
MS, TY and NM participated in the study conception and design, data collection, data analysis, reviewed the paper and approved the final draft for submission. TY and NM participated in the study conception and design, funding acquisition, data collection, data analysis, reviewed the paper and approved the final draft for submission. RH participated in data collection, reviewed the paper and approved the final draft for submission. KS, NG and SY participated in the study data collection, funding acquisition, data analysis, reviewed the paper. YM, YS, YO, YG, HH, MY, YY, K Nakagawa, K Naoki, TT, TK, NY, SW and YO participated in data collection, reviewed the paper and approved the final draft for submission.
Corresponding author
Ethics declarations
Competing interests
Dr. TY reports grants from Takeda, Daiichi-Sankyo, AbbVie, and Bristol-Myers Squibb; grants and personal fees from Ono, Novartis, Merck Sharp & Dohme, and Amgen; and personal fees from AstraZeneca. KK reports personal fees from Chugai, ArcherDX, Eli Lilly, Roche, and Taiho outside the submitted work. Dr. YM reports grants from Grant-in-Aid for Scientific Research on Innovative Areas, Hitachi, Ltd., the National Cancer Center Research and Development Fund, and Olympus, and personal fees from AstraZeneca. Drs. MS, KS, NG, SY, TI, KM, MY, YY, K Nakagawa, and S-iW declare no competing interests. Dr. YS reports grants from Janssen and Japan Clinical Research Operations. KK reports personal fees from AstraZeneca, Bristol-Myers Squibb, Chugai, Eli Lilly, and Ono outside the submitted work. Dr. YO reports grants from AbbVie, grants from Roche, and personal fees from Bristol-Myers Squibb Grants, Nippon Boehringer Ingelheim, and AstraZeneca. KK reports personal fees from Chugai, Eli Lilly, Ono Pharma Co. Ltd., Taiho, and Pfizer Taiho Pharma Co. Ltd., outside the submitted work. Dr. YO reports grants from AbbVie, Bristol-Myers Squibb Grants, Kyorin, and Preferred Network; and grants and personal fees from AZK, Daiichi-Sankyo, Eli Lilly, Ono, Novartis, and Pfizer; and personal fees from AstraZeneca. KK reports personal fees from Boehringer Ingelheim, Chugai, Guardant Health Inc., Illumina, Merck Sharp & Dohme, Taiho, and Thermo Fisher outside the submitted work. Dr. HH reports grants and personal fees from Chugai, Ono, and Roche; grants from AbbVie, Bristol-Myers Squibb, Daiichi-Sankyo, Genomic Health, Janssen, and Merck Biopharma, and personal fees from AstraZeneca. KK reports personal fees from Merck Sharp & Dohme, Novartis, Eli Lilly, and Kyowa-Kirin, outside the submitted work. Dr. K Naoki reports personal fees from AstraZeneca, Chugai Pharmaceutical, Bristol-Myers Squibb, and Nippon Boehringer Ingelheim outside the submitted work. Dr. TT reports grants from Japan Agency for Medical Research and Development, and Foundation for Promotion Cancer Research Grant for Medical Research, and personal fees from Nippon Medical School Foundation, outside the submitted work. Dr. RH reports grants from JST AIP-PRISM (Grant Number JPMJCR18Y4), from null, outside the submitted work. Dr. Yamamoto reports grants from Chugai, Taiho, Eisai, Lilly, Quintiles, Astellas, BMS, Novartis, Daiichi-Sankyo, Pfizer, Boehringer Ingelheim, Kyowa-Hakko Kirin, Bayer, ONO PHARMACEUTICAL CO., LTD, and Takeda; and personal fees from ONO PHARMACEUTICAL CO., LTD, Chugai, Eisai, Boehringer Ingelheim, and Cmic; and grants from Janssen Pharma, MSD, Merck, GSK, Sumitomo Dainippon, Chiome Bioscience Inc., Otsuka, Carna Biosciences, Genmab, TORAY, KAKEN, Shionogi, AstraZeneca, and Cmic; and personal fees from Eisai, Daiichi-Sankyo, MERCK, and Healios, outside the submitted work. Dr. RH reports grants from NEC and Ono, and personal fees from AstraZeneca, Chugai, MSD, Novartis, Roche, Daiichi-Sankyo, Takeda, Eli Lilly, and Janssen, outside the submitted work. Dr. TK reports grants from AMED, outside the submitted work. Dr. YO reports grants from Taiho, Eli Lilly, Kyorin, Daiichi-Sankyo, Dainippon-Sumitomo, Janssen, Kissei, LOXO, Novartis, and Takeda, and personal fees from AstraZeneca. KK reports personal fees from Amgen, AnHeeart Therapeutics Inc., Bayer, Bristol-Myers Squibb, Celltrion, Chugai, Kyowa-Hakko Kirin, Merck Sharp & Dohme, Nippon Kayaku, Ono Pharmaceutical Co., Ltd., Pfizer, Taiho, and Eli Lilly, outside the submitted work.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of the National Cancer Center Hospital (2005-109, 2019-123). The written informed consent was obtained from each patient through the comprehensive consent form in the facilities. The study was performed in accordance with the Declaration of Helsinki.
Consent for publication
Not applicable.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: The original online version of this article was revised: In the caption of fig 3d is typo. The sentence “The top box indicates the DLL3 expression (z score). Heatmaps: expression of 18-gene-γ-related T-cell gene expression profiles (T-cell GEPs) and 13 chemokine markers of high and low DLL3 expression tumors are shown.” should read “The top box indicates the DLL3 expression (z score). Heatmaps: expression of 18-gene-γ-related T-cell gene expression profiles (T-cell GEPs) and 12 chemokine markers of high and low DLL3 expression tumors are shown”.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shirasawa, M., Yoshida, T., Shiraishi, K. et al. Tumor microenvironment-mediated immune profiles and efficacy of anti-PD-L1 antibody plus chemotherapy stratified by DLL3 expression in small-cell lung cancer. Br J Cancer 129, 2003–2013 (2023). https://doi.org/10.1038/s41416-023-02427-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-023-02427-3
This article is cited by
-
Prognostic significance of immunohistochemical classification utilizing biopsy specimens in patients with extensive-disease small-cell lung cancer treated with first-line chemotherapy and immune checkpoint inhibitors
Journal of Cancer Research and Clinical Oncology (2024)