Abstract
Background
Early detection of BCR::ABL1-like ALL could impact treatment management and improve the overall survival/outcome. BCR::ABL1-like ALL cases are characterised by diverse genetic alterations activating cytokine receptors and kinase signalling. Its detection is still an unmet need in low–middle-income countries due to the unavailability of a patented TLDA assay.
Methods
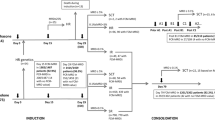
This study’s rationale is to identify BCR::ABL1-like ALLs using the PHi-RACE classifier, followed by the characterisation of underlying adverse genetic alterations in recurrent gene abnormalities negative (RGAneg) B-ALLs (n = 108).
Results
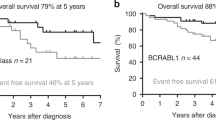
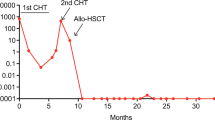
We identified 34.25% (37/108) BCR::ABL1-like ALLs using PHi-RACE classifier, characterised by TSLPR/CRLF2 expression (11.58%), IKZF1 (Δ4–7) deletion (18.9%) and chimeric gene fusions (34.61%). In overexpressed TSLPR/CRLF2 BCR::ABL1-like ALLs, we identified 33.33% (1/3) CRLF2::IGH and 33.33% (1/3) EPOR::IGH rearrangements with concomitant JAK2 mutation R683S (50%). We identified 18.91% CD13 (P = 0.02) and 27.02% CD33 (P = 0.05) aberrant myeloid markers positivity, which was significantly higher in BCR::ABL1-like ALLs compared to non-BCR::ABL1-like ALLs. MRD positivity was considerably higher (40% in BCR::ABL1-like vs. 19.29% in non-BCR::ABL1-like ALLs).
Conclusions
With this practical approach, we reported a high incidence of BCR::ABL1-like ALLs, and a lower frequency of CRLF2 alteration & associated CGFs. Recognising this entity, early at diagnosis is crucial to optimise personalised treatment strategies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All research data generated during this study are included in this research article.
References
Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–405.
Alaggio R, Amador C, Anagnostopoulos I, Attygalle AD, Araujo IBO, Berti E, et al. The 5th edition of the World Health Organization Classification of haematolymphoid tumours: lymphoid neoplasms. Leukemia. 2022;36:1720–48.
Mullighan CG, Su X, Zhang J, Radtke I, Phillips LA, Miller CB, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009;360:470–80.
Den Boer ML, van Slegtenhorst M, De Menezes RX, Cheok MH, Buijs-Gladdines JG, Peters ST, et al. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. Lancet Oncol. 2009;10:125–34.
Roberts KG, Li Y, Payne-Turner D, Harvey RC, Yang YL, Pei D, et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med. 2014;371:1005–15.
Mullighan CG. The genomic landscape of acute lymphoblastic leukemia in children and young adults. Hematol Am Soc Hematol Educ Program. 2014;2014:174–80.
Roberts KG, Gu Z, Payne-Turner D, McCastlain K, Harvey RC, Chen IM, et al. High frequency and poor outcome of Philadelphia chromosome-like acute lymphoblastic leukemia in adults. J Clin Oncol. 2017;35:394–401.
Reshmi SC, Harvey RC, Roberts KG, Stonerock E, Smith A, Jenkins H, et al. Targetable kinase gene fusions in high-risk B-ALL: a study from the Children’s Oncology Group. Blood. 2017;129:3352–61.
Ofran Y, Izraeli S. BCR-ABL (Ph)-like acute leukemia-pathogenesis, diagnosis and therapeutic options. Blood Rev. 2017;31:11–6.
Jain N, Roberts KG, Jabbour E, Patel K, Eterovic AK, Chen K, et al. Ph-like acute lymphoblastic leukemia: a high-risk subtype in adults. Blood. 2017;129:572–81.
Herold T, Schneider S, Metzeler KH, Neumann M, Hartmann L, Roberts KG, et al. Adults with Philadelphia chromosome-like acute lymphoblastic leukemia frequently have IGH-CRLF2 and JAK2 mutations, persistence of minimal residual disease and poor prognosis. Haematologica. 2017;102:130–8.
Heatley SL, Sadras T, Kok CH, Nievergall E, Quek K, Dang P, et al. High prevalence of relapse in children with Philadelphia-like acute lymphoblastic leukemia despite risk-adapted treatment. Haematologica. 2017;102:e490–e3.
Wells J, Jain N, Konopleva M. Philadelphia chromosome-like acute lymphoblastic leukemia: progress in a new cancer subtype. Clin Adv Hematol Oncol. 2017;15:554–61.
Tasian SK, Loh ML, Hunger SP. Philadelphia chromosome-like acute lymphoblastic leukemia. Blood. 2017;130:2064–72.
Roberts KG, Reshmi SC, Harvey RC, Chen IM, Patel K, Stonerock E, et al. Genomic and outcome analyses of Ph-like ALL in NCI standard-risk patients: a report from the Children’s Oncology Group. Blood. 2018;132:815–24.
Chiaretti S, Messina M, Grammatico S, Piciocchi A, Fedullo AL, Di Giacomo F, et al. Rapid identification of BCR/ABL1-like acute lymphoblastic leukaemia patients using a predictive statistical model based on quantitative real time-polymerase chain reaction: clinical, prognostic and therapeutic implications. Br J Haematol. 2018;181:642–52.
Siegele BJ, Nardi V. Laboratory testing in BCR-ABL1-like (Philadelphia-like) B-lymphoblastic leukemia/lymphoma. Am J Hematol. 2018;93:971–7.
Chiaretti S, Messina M, Foa R. BCR/ABL1-like acute lymphoblastic leukemia: how to diagnose and treat? Cancer. 2019;125:194–204.
Shiraz P, Payne KJ, Muffly L. The current genomic and molecular landscape of Philadelphia-like acute lymphoblastic leukemia. Int J Mol Sci. 2020;21:2193.
Jain S, Abraham A. BCR-ABL1-like B-acute lymphoblastic leukemia/lymphoma: a comprehensive review. Arch Pathol Lab Med. 2020;144:150–5.
Ribera JM. Philadelphia chromosome-like acute lymphoblastic leukemia. Still a pending matter. Haematologica. 2021;106:1514–6.
Chiaretti S, Messina M, Della Starza I, Piciocchi A, Cafforio L, Cavalli M, et al. Philadelphia-like acute lymphoblastic leukemia is associated with minimal residual disease persistence and poor outcome. First report of the minimal residual disease-oriented GIMEMA LAL1913. Haematologica. 2021;106:1559–68.
Gupta DG, Varma N, Kumar A, Naseem S, Sachdeva MUS, Binota J, et al. PHi-RACE: PGIMER in-house rapid & cost effective classifier for the detection of BCR-ABL1-like acute lymphoblastic leukaemia in Indian patients. Leuk Lymphoma. 2022;63:633–43.
Plotka A, Lewandowski K. BCR/ABL1-like acute lymphoblastic leukemia: from diagnostic approaches to molecularly targeted therapy. Acta Haematol. 2022;145:122–31.
Harvey RC, Mullighan CG, Chen IM, Wharton W, Mikhail FM, Carroll AJ, et al. Rearrangement of CRLF2 is associated with mutation of JAK kinases, alteration of IKZF1, Hispanic/Latino ethnicity, and a poor outcome in pediatric B-progenitor acute lymphoblastic leukemia. Blood. 2010;115:5312–21.
Chiaretti S, Gianfelici V, O’Brien SM, Mullighan CG. Advances in the genetics and therapy of acute lymphoblastic leukemia. Am Soc Clin Oncol Educ Book. 2016;35:e314–22.
Totadri S, Singh M, Trehan A, Varma N, Bhatia P. Keeping PACE with Ph positive to Ph-like detection in B-lineage acute lymphoblastic leukemia: a practical and cost effective (PACE) approach in a resource constrained setting. Indian J Hematol Blood Transfus. 2018;34:595–601.
Sanchez R, Ribera J, Morgades M, Ayala R, Onecha E, Ruiz-Heredia Y, et al. A novel targeted RNA-Seq panel identifies a subset of adult patients with acute lymphoblastic leukemia with BCR-ABL1-like characteristics. Blood Cancer J. 2020;10:43.
Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453:110–4.
Harvey RC, Kang H, Roberts KG, Chen I-ML, Atlas SR, Bedrick EJ, et al. Development and validation of a highly sensitive and specific gene expression classifier to prospectively screen and identify B-precursor acute lymphoblastic leukemia (ALL) patients with a Philadelphia chromosome-like (“Ph-like” or “BCR-ABL1-Like”) signature for therapeutic targeting and clinical intervention. Blood. 2013;122:826.
Boer JM, Koenders JE, van der Holt B, Exalto C, Sanders MA, Cornelissen JJ, et al. Expression profiling of adult acute lymphoblastic leukemia identifies a BCR-ABL1-like subgroup characterized by high non-response and relapse rates. Haematologica. 2015;100:e261–4.
Chiaretti S, Brugnoletti F, Messina M, Paoloni F, Fedullo AL, Piciocchi A, et al. CRLF2 overexpression identifies an unfavourable subgroup of adult B-cell precursor acute lymphoblastic leukemia lacking recurrent genetic abnormalities. Leuk Res. 2016;41:36–42.
Tsaur G, Muhacheva T, Kovalev S, Druy A, Sibiryakov P, Olshanskaya Y, et al. Application of real-time PCR for the detection of BCR-ABL1-like group in pediatric acute lymphoblastic leukemia patients. Blood. 2018;132:1376.
Fasan Annette KW, Nadarajah N, Weber S, Schindela S, Schlenther N, Schnittger S, et al. Three steps to the diagnosis of adult Ph-like ALL. Blood. 2015;126:2610.
Patkar N, Bhanshe P, Rajpal S, Joshi S, Chaudhary S, Chatterjee G, et al. NARASIMHA: novel assay based on targeted rna sequencing to identify chimeric gene fusions in hematological malignancies. Blood Cancer J 2020;10:50.
Virk H, Rana S, Sharma P, Bose PL, Yadav DD, Sachdeva MUS, et al. Hematological characteristics, cytogenetic features, and post-induction measurable residual disease in thymic stromal lymphopoietin receptor (TSLPR) overexpressed B-cell acute lymphoblastic leukemia in an Indian cohort. Ann Hematol. 2021;100:2031–41.
Sharma P, Rana S, Virk H, Sachdeva MUS, Sharma P, Varma N, et al. The frequency, hematological characteristics, and end-of induction residual disease in B-acute lymphoblastic leukemia with BCR-ABL1-like chimeric gene fusions in a high-risk cohort from India. Leuk Lymphoma. 2021;63:2474–8.
Gupta DG, Varma N, Naseem S, Sachdeva MUS, Bose P, Binota J, et al. Characterization of immunophenotypic aberrancies with respect to common fusion transcripts in B-cell precursor acute lymphoblastic leukemia: a report of 986 Indian patients. Turk J Haematol. 2022;39:1–12.
Sharma M, Sachdeva MU, Varma N, Varma S, Marwaha RK. Characterization of immunophenotypic aberrancies in adult and childhood acute lymphoblastic leukemia: a study from Northern India. J Cancer Res Ther. 2016;12:620–6.
Malhotra P, Varma S, Varma N, Kumari S, Das R, Jain S, et al. Outcome of adult acute lymphoblastic leukemia with BFM protocol in a resource-constrained setting. Leuk Lymphoma. 2007;48:1173–8.
Bommannan K, Singh Sachdeva MU, Varma N, Bose P, Bansal D. Role of mid-induction peripheral blood minimal residual disease detection in pediatric B-lineage acute lymphoblastic leukemia. Indian Pediatr. 2016;53:1065–8.
Tembhare PR, Subramanian PgPG, Ghogale S, Chatterjee G, Patkar NV, Gupta A, et al. A high-sensitivity 10-color flow cytometric minimal residual disease assay in B-lymphoblastic leukemia/lymphoma can easily achieve the sensitivity of 2-in-10(6) and is superior to standard minimal residual disease assay: a study of 622 patients. Cytom B Clin Cytom. 2020;98:57–67.
Stow P, Key L, Chen X, Pan Q, Neale GA, Coustan-Smith E, et al. Clinical significance of low levels of minimal residual disease at the end of remission induction therapy in childhood acute lymphoblastic leukemia. Blood. 2010;115:4657–63.
Pakakasama S, Kajanachumpol S, Kanjanapongkul S, Sirachainan N, Meekaewkunchorn A, Ningsanond V, et al. Simple multiplex RT-PCR for identifying common fusion transcripts in childhood acute leukemia. Int J Lab Hematol. 2008;30:286–91.
Bhatia P, Binota J, Varma N, Bansal D, Trehan A, Marwaha RK, et al. Incidence of common chimeric fusion transcripts in B-cell acute lymphoblastic leukemia: an Indian perspective. Acta Haematol. 2012;128:17–9.
Sharma P, Rana S, Sreedharanunni S, Gautam A, Sachdeva MUS, Naseem S, et al. An evaluation of a fluorescence in situ hybridization strategy using air-dried blood and bone-marrow smears in the risk stratification of pediatric B-lineage acute lymphoblastic leukemia in resource-limited settings. J Pediatr Hematol Oncol. 2021;43:e481–e5.
Bugarin C, Sarno J, Palmi C, Savino AM, te Kronnie G, Dworzak M, et al. Fine tuning of surface CRLF2 expression and its associated signaling profile in childhood B-cell precursor acute lymphoblastic leukemia. Haematologica. 2015;100:e229–32.
Boer JM, van der Veer A, Rizopoulos D, Fiocco M, Sonneveld E, de Groot-Kruseman HA, et al. Prognostic value of rare IKZF1 deletion in childhood B-cell precursor acute lymphoblastic leukemia: an international collaborative study. Leukemia. 2016;30:32–8.
Hashiguchi J, Onozawa M, Okada K, Amano T, Hatanaka KC, Nishihara H, et al. Quantitative detection of IKZF1 deletion by digital PCR in patients with acute lymphoblastic leukemia. Int J Lab Hematol. 2019;41:e38–e40.
Kastner P, Dupuis A, Gaub MP, Herbrecht R, Lutz P, Chan S. Function of Ikaros as a tumor suppressor in B cell acute lymphoblastic leukemia. Am J Blood Res. 2013;3:1–13.
Iacobucci I, Storlazzi CT, Cilloni D, Lonetti A, Ottaviani E, Soverini S, et al. Identification and molecular characterization of recurrent genomic deletions on 7p12 in the IKZF1 gene in a large cohort of BCR-ABL1-positive acute lymphoblastic leukemia patients: on behalf of Gruppo Italiano Malattie Ematologiche dell’Adulto Acute Leukemia Working Party (GIMEMA AL WP). Blood. 2009;114:2159–67.
Iacobucci I, Iraci N, Messina M, Lonetti A, Chiaretti S, Valli E, et al. IKAROS deletions dictate a unique gene expression signature in patients with adult B-cell acute lymphoblastic leukemia. PLoS ONE. 2012;7:e40934.
Kuiper RP, Waanders E, van der Velden VH, van Reijmersdal SV, Venkatachalam R, Scheijen B, et al. IKZF1 deletions predict relapse in uniformly treated pediatric precursor B-ALL. Leukemia. 2010;24:1258–64.
Yamashita Y, Shimada A, Yamada T, Yamaji K, Hori T, Tsurusawa M, et al. IKZF1 and CRLF2 gene alterations correlate with poor prognosis in Japanese BCR-ABL1-negative high-risk B-cell precursor acute lymphoblastic leukemia. Pediatr Blood Cancer. 2013;60:1587–92.
Martinelli G, Iacobucci I, Storlazzi CT, Vignetti M, Paoloni F, Cilloni D, et al. IKZF1 (Ikaros) deletions in BCR-ABL1-positive acute lymphoblastic leukemia are associated with short disease-free survival and high rate of cumulative incidence of relapse: a GIMEMA AL WP report. J Clin Oncol. 2009;27:5202–7.
Caye A, Beldjord K, Mass-Malo K, Drunat S, Soulier J, Gandemer V, et al. Breakpoint-specific multiplex polymerase chain reaction allows the detection of IKZF1 intragenic deletions and minimal residual disease monitoring in B-cell precursor acute lymphoblastic leukemia. Haematologica. 2013;98:597–601.
Mi JQ, Wang X, Yao Y, Lu HJ, Jiang XX, Zhou JF, et al. Newly diagnosed acute lymphoblastic leukemia in China (II): prognosis related to genetic abnormalities in a series of 1091 cases. Leukemia. 2012;26:1507–16.
Palmi C, Vendramini E, Silvestri D, Longinotti G, Frison D, Cario G, et al. Poor prognosis for P2RY8-CRLF2 fusion but not for CRLF2 over-expression in children with intermediate risk B-cell precursor acute lymphoblastic leukemia. Leukemia. 2012;26:2245–53.
Liu X, Zhang L, Zou Y, Chang L, Wei W, Ruan M, et al. [Significance of ikaros family zinc finger 1 deletion in pediatric B-acute lymphoblastic leukemia without reproducible cytogenetic abnormalities]. Zhonghua Er Ke Za Zhi. 2016;54:126–30.
Asai D, Imamura T, Suenobu S, Saito A, Hasegawa D, Deguchi T, et al. IKZF1 deletion is associated with a poor outcome in pediatric B-cell precursor acute lymphoblastic leukemia in Japan. Cancer Med. 2013;2:412–9.
Yoda A, Yoda Y, Chiaretti S, Bar-Natan M, Mani K, Rodig SJ, et al. Functional screening identifies CRLF2 in precursor B-cell acute lymphoblastic leukemia. Proc Natl Acad Sci USA. 2010;107:252–7.
Chen IM, Harvey RC, Mullighan CG, Gastier-Foster J, Wharton W, Kang H, et al. Outcome modeling with CRLF2, IKZF1, JAK, and minimal residual disease in pediatric acute lymphoblastic leukemia: a Children’s Oncology Group study. Blood. 2012;119:3512–22.
Cario G, Zimmermann M, Romey R, Gesk S, Vater I, Harbott J, et al. Presence of the P2RY8-CRLF2 rearrangement is associated with a poor prognosis in non-high-risk precursor B-cell acute lymphoblastic leukemia in children treated according to the ALL-BFM 2000 protocol. Blood. 2010;115:5393–7.
Pastorczak A, Sedek L, Braun M, Madzio J, Sonsala A, Twardoch M, et al. Surface expression of cytokine receptor-like factor 2 increases risk of relapse in pediatric acute lymphoblastic leukemia patients harboring IKZF1 deletions. Oncotarget. 2018;9:25971–82.
Tasian SK, Dai Y, Devidas M, Roberts KG, Harvey RC, Chen I-ML, et al. Outcomes of patients with CRLF2-overexpressing acute lymphoblastic leukemia without down syndrome: a report from the Children’s Oncology Group. Blood. 2020;136:45–6.
Acknowledgements
I sincerely thank the faculty of Haematology, technical staff, and clerical staff, project staff (Mrs Sonia) for providing valuable support for this study. I would like to extend my sincere thanks to Dr. Praveen (PGIMER), Dr Pramod Singh (PGIMER), Dr Mathew Li Arwood (Ann Roberts H. Lurie Children’s Hospital), and Dr Loretta S Li (Ann Roberts H. Lurie Children’s Hospital) for their valuable suggestions in this manuscript.
Funding
This research study was supported by the Intramural Grant provided by PGIMER, Chandigarh, India (No.71/2-Edu-16/937/18/03/2019). Dr. Dikshat Gopal Gupta received financial assistance from the Indian Council of Medical Education and Research (ICMR), New Delhi, India.
Author information
Authors and Affiliations
Contributions
DGG and NV designed the experiments and analysed the generated data; DGG, SS, JB and PB performed the experiments. DGG and NV wrote and prepared the manuscript; PM, AK and SV provided the adult B-ALL samples. SAA read, reviewed and helped us properly restructure the manuscript. All other authors read, provided intellectual comments, and approved the submitted manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
PGIMER constituted Institutional Ethics Committee had approved this research study (vide no. INT/IEC/2019/000611 dated March 19, 2019). In all, 2–3 ml of aspirated bone marrow (BM) or 3–5 ml of peripheral blood (PB) samples were collected, after obtaining the informed consent.
Consent for publication
Not applicable.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gupta, D.G., Varma, N., Sreedharanunni, S. et al. ‘Evaluation of adverse prognostic gene alterations & MRD positivity in BCR::ABL1-like B-lineage acute lymphoblastic leukaemia patients, in a resource-constrained setting. Br J Cancer 129, 143–152 (2023). https://doi.org/10.1038/s41416-023-02294-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-023-02294-y